|
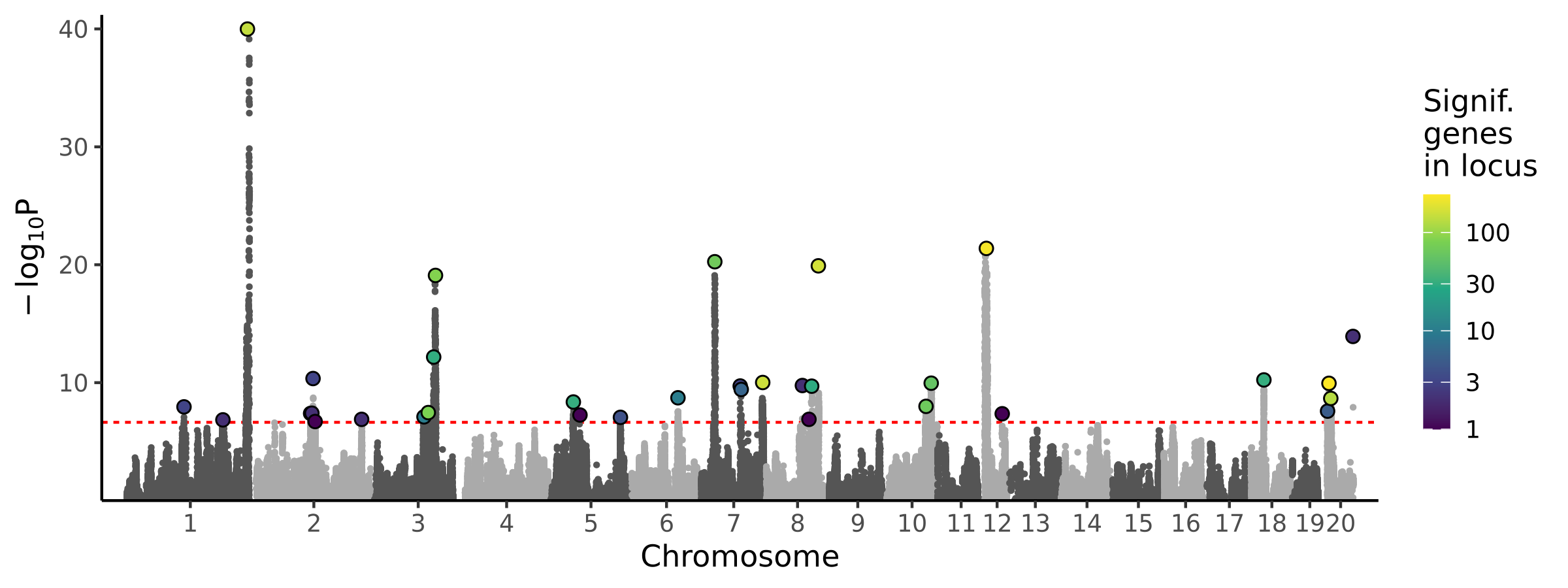
BMI with tail
|
11 |
135 |
90 |
83.3 |
1 |
0.00e+00 |
Abca6
Abca9
Abhd18
Ablim1
Acsl5
Adrb1
Afap1l2
Amz2
Ankrd50
Anxa5
Apc
Apoh
Arsg
Atp11b
Atp1b3
Atrnl1
Axin2
Bbs7
Brd8
Cacul1
Casp7
Ccdc186
Cd99
Cdc23
Cep112
Ces2c
Chst2
Dclre1a
Dcun1d1
Dennd10
Dstn
Dtd1
Dzank1
Ehd4
Emx2os
Eno4
Epb41l4a
Ext1
Fam13b
Fam204a
Fam20a
Fat4
Fermt3
Fhip2a
Flrt3
Gfra1
Gk5
Gpam
Grk5
Gucy2g
Habp2
Hdac7
Hmgcll1
Hspa12a
Jade1
Kif16b
Kif20a
Klhdc8b
Larp1b
LOC100912202
LOC102547573
LOC102548155
LOC102548513
LOC102548590
LOC102550112
LOC102550304
LOC102550729
LOC102551125
LOC102552777
LOC103694460
LOC103694952
LOC120095201
LOC120098271
LOC120100065
LOC120100068
LOC120101717
LOC120101719
Macrod2
Map2k6
Mapkbp1
Mccc1
Med30
Nanos1
Ndufaf5
Nhlrc2
Nme5
Nrap
Otor
PCOLCE2
Pcsk2
Pfkm
Pkd2l2
Pla2g4b
Plekhs1
Pls1
Pnlip
Polr3f
Prdx3
Prkca
Prlhr
Qrfpr
Rab11fip2
Rasa2
Rbbp9
Reep5
RGD1565459
Rgs9
Rnf7
Rpap3
Rps6ka4
Rrbp1
Scg3
Sec23b
Sfxn4
Slc16a6
Smim26
Snrpb2
Sptbn5
Srp19
Tasp1
Tcf7l2
Tectb
Tfdp2
Tmcc3
Tmem106c
Trpc1
Trps1
Trub1
U2surp
Vti1a
Wipi1
Zdhhc6
Zfp133
Zfp267
Zfp950
|
|
BMI without tail
|
15.75 |
32 |
16 |
14.8 |
1 |
4.63e-170 |
Cacul1
Ces2c
Dennd10
Dstn
Emx2os
Epb41l4a
Fam204a
Grk5
Hspa12a
Kif16b
Klhdc8b
LOC102548590
LOC102550729
LOC120100068
LOC120101717
LOC120101719
Mir331
Nanos1
Otor
Pcsk2
Prdx3
Prlhr
Rab11fip2
Rnf7
Rrbp1
Sec23b
Sfxn4
Snrpb2
Tfdp2
Trpc1
Trps1
Zfp950
|
|
Epididymis fat weight
|
16.95 |
77 |
48 |
44.4 |
0.99 |
0.00e+00 |
Abca6
Abca9
Ablim1
Acsl5
Add3
Adrb1
Afap1l2
Amz2
Apoh
Arsg
Atrnl1
Axin2
Cacul1
Casp7
Ccdc186
Cep112
Ces2c
Dclre1a
Dennd10
Dstn
Emx2os
Eno4
Fam204a
Fam20a
Fhip2a
Flrt3
Gfra1
Gpam
Grk5
Gucy2g
Habp2
Hspa12a
Ism1
Kif16b
LOC100912202
LOC102547573
LOC102548513
LOC102550112
LOC102550729
LOC102551125
LOC120095201
LOC120100064
LOC120100065
LOC120100068
LOC120101717
LOC120101719
Macrod2
Mxi1
Nanos1
Nhlrc2
Nrap
Otor
Pcsk2
Plekhs1
Pnlip
Prdx3
Prkar1a
Prlhr
Rab11fip2
Rbbp9
RGD1565459
Rgs9
Rrbp1
Sec23b
Sfxn4
Shoc2
Slc16a6
Snrpb2
Sptlc3
Tasp1
Tcf7l2
Tectb
Trub1
Vti1a
Wipi1
Zdhhc6
Zfp950
|
|
Heart weight
|
17.24 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Left kidney weight
|
10.37 |
73 |
43 |
39.8 |
0.98 |
0.00e+00 |
Acsl5
Arhgef18
Asap3
Brca2
Cacul1
Camsap3
Casp7
Cd209a
Cd99
Cers4
Ces2c
Dennd10
Elavl1
Emx2os
Fam204a
Fry
Gpam
Grk5
Gucy2g
Hmgb1
Hmgcl
Hnrnpr
Hsph1
Insr
Katnal1
Kl
Klhdc8b
Lnc001
LOC102546827
LOC102548389
LOC102549089
LOC102549494
LOC102550729
LOC102552452
LOC102553253
LOC103690858
LOC103694460
LOC108352411
LOC120095826
LOC120095830
LOC120095870
LOC120095871
LOC120095873
LOC120100064
LOC120100068
Map2k7
Mcemp1
Mcoln1
Medag
N4bp2l1
N4bp2l2
Nanos1
Pcp2
Pet100
Pex11g
Pi16
Pnpla6
Prdx3
Prlhr
Rab11fip2
Rxfp2
Sfxn4
Snapc2
Stard13
Stxbp2
Tcf7l2
Timm44
Trappc5
Vti1a
Wdr95
Xab2
Zfp950
Zfp958
|
|
Right kidney weight
|
9.33 |
80 |
43 |
39.8 |
0.99 |
0.00e+00 |
Ablim1
Acsl5
Adrb1
Arhgef18
Atrnl1
B3glct
Brca2
Cacul1
Camsap3
Casp7
Cd209a
Cers4
Ces2c
Dennd10
Elavl1
Emx2os
Fam204a
Fhip2a
Fry
Gpam
Grk5
Gucy2g
Habp2
Hmgb1
Hsph1
Insr
Katnal1
Kl
Klhdc8b
Lnc001
LOC102546827
LOC102547573
LOC102548389
LOC102549089
LOC102549494
LOC102550729
LOC102552777
LOC102553253
LOC102554603
LOC103690858
LOC103694460
LOC108352411
LOC120095826
LOC120095830
LOC120095870
LOC120095871
LOC120095873
LOC120095879
LOC120100064
LOC120100065
LOC120100068
Map2k7
Mcemp1
Mcoln1
Medag
N4bp2l1
N4bp2l2
Nanos1
Nhlrc2
Nrap
Pcp2
Pet100
Pex11g
Plekhs1
Prdx3
Prlhr
Rab11fip2
Rxfp2
Sfxn4
Snapc2
Stard13
Stxbp2
Tcf7l2
Trappc5
Uspl1
Vti1a
Wdr95
Xab2
Zfp950
Zfp958
|
|
Tail length
|
8.22 |
125 |
47 |
43.5 |
0.99 |
0.00e+00 |
Abcf1
Als2cl
Amdhd1
Arhgef18
Arih2
Atrip
Bag6
Brca2
Camp
Camsap3
Ccdc12
Ccdc38
Ccdc51
Cchcr1
Cd209a
Cdc25a
Cdk17
Cep83
Cers4
Cspg5
Dclk3
Ddr1
Dhx16
Elavl1
Elk3
Elp6
Epm2aip1
Fam240a
Fgd6
Fry
Gnl1
Gtf2h4
Insr
Kif9
Kl
Klhdc8b
Klhl18
Lnc001
LOC102548389
LOC102548950
LOC102549089
LOC102552452
LOC102552504
LOC102553539
LOC102554603
LOC102556485
LOC103690858
LOC108351781
LOC120093125
LOC120093556
LOC120093559
LOC120093563
LOC120095826
LOC120095871
LOC120095873
LOC120098967
Lrrc2
Lrrfip2
Lta4h
Map2k7
Map4
Mcoln1
Metap2
Mir331
Mlh1
Mrpl42
Mrps18b
Myl3
N4bp2l1
N4bp2l2
Nbeal2
Nckipsd
Ndufa12
Ngp
Nme6
Nr2c1
Nradd
Nrm
Ntn4
Nudt4
Pcp2
Pet100
Pex11g
Pfkfb4
Plxnb1
Plxnc1
Pnpla6
Prr3
Prss50
Pth1r
Ptpn23
RT1-CE1
RT1-CE10
RT1-CE11
RT1-CE14
RT1-CE15
RT1-N2
RT1-N3
RT1-S3
RT1-T18
RT1-T24-1
RT1-T24-2
RT1-T24-4
Rtp3
Scap
Setd2
Shisa5
Smarcc1
Snapc2
Snrpf
Socs2
Stard13
Stxbp2
Tcf19
Timm44
Tmcc3
Tmie
Trank1
Trappc5
Trex1
Ube2n
Vars2
Vezt
Xab2
Zfp958
|
|
Length with tail
|
10.54 |
177 |
131 |
121.3 |
0.99 |
0.00e+00 |
Abcf1
Als2cl
Amdhd1
Arhgef18
Arih2
Arl5c
Atat1
Atp6v1d
Atrip
B3glct
Bag6
Brca2
Cacnb1
Camp
Camsap3
Cb707485
Ccdc12
Ccdc38
Ccdc51
Cchcr1
Cd209a
Cdc25a
Cdk17
Cdsn
Cep83
Cep83os
Cers4
Cisd3
Cspg5
Ctxn1
Cwc25
Dclk3
Ddr1
Dhx16
Ebf4
Elavl1
Elk3
Elp6
Epm2aip1
Epop
Fam240a
Fbxl20
Fgd6
Fkbp5
Flot1
Fry
G4
Gnl1
Gtf2h4
Hmgb1
Hsph1
Insr
Katnal1
Kif16b
Kif9
Kl
Klhdc8b
Klhl18
Lnc001
LOC102546827
LOC102548260
LOC102548389
LOC102548848
LOC102548950
LOC102549089
LOC102549494
LOC102552452
LOC102552504
LOC102553244
LOC102553253
LOC102553539
LOC102554603
LOC102556485
LOC103690858
LOC108349027
LOC108351781
LOC108352411
LOC120093125
LOC120093556
LOC120093559
LOC120093563
LOC120095826
LOC120095830
LOC120095870
LOC120095871
LOC120095873
LOC120098966
LOC120098967
Lrrc2
Lrrc8e
Lrrfip2
Lta4h
Map2k7
Map4
Mcemp1
Mcoln1
Medag
Metap2
Mir331
Mlh1
Mllt6
Mrpl42
Mrps18b
Myl3
N4bp2l1
N4bp2l2
Nbeal2
Nckipsd
Ndufa12
Ngp
Nme6
Nr2c1
Nradd
Nrm
Ntn4
Nudt4
Otor
Pced1a
Pcp2
Pds5b
Pdyn
Pet100
Pex11g
Pfkfb4
Plxnb1
Plxnc1
Pnpla6
Prr3
Prss50
Pth1r
Ptpn23
Ptpra
Retn
Rpl19
RT1-CE10
RT1-CE11
RT1-CE4
RT1-N2
RT1-N3
RT1-O1
RT1-S2
RT1-S3
RT1-T18
RT1-T24-1
RT1-T24-2
RT1-T24-3
RT1-T24-4
Rtp3
Rxfp2
Scap
Setd2
Shisa5
Slc26a8
Smarcc1
Snapc2
Snrpb
Snrpb2
Snrpf
Socs2
Srcin1
Stac2
Stard13
Stxbp2
Tcf19
Timm44
Tma7
Tmcc3
Tmie
Trank1
Trappc5
Trex1
Ube2n
Vars2
Vezt
Wdr95
Xab2
Zfp958
|
|
Length without tail
|
11.8 |
73 |
34 |
31.5 |
0.99 |
0.00e+00 |
Alox5ap
Arhgef18
Asb8
Brca2
Cacul1
Camsap3
Cd209a
Cers4
Dclk3
Ddx23
Elavl1
Fgd6
Fry
Grk5
Hdac7
Hmgb1
Hsph1
Insr
Kansl2
Katnal1
Kl
Lnc001
LOC102546827
LOC102548155
LOC102548389
LOC102549089
LOC102549494
LOC102552452
LOC102553253
LOC102553539
LOC103690858
LOC108351548
LOC108352411
LOC120093563
LOC120095826
LOC120095830
LOC120095871
LOC120095873
LOC120095879
Lrrc8e
Lta4h
Map2k7
Mcemp1
Mcoln1
Medag
Mir331
Mrpl42
N4bp2l1
N4bp2l2
Ndufa12
Nr2c1
Or8s2
Pcp2
Pet100
Pex11g
Pfkm
Plxnc1
Prlhr
Rapgef3
Rpap3
Rxfp2
Slc48a1
Snapc2
Stard13
Stxbp2
Timm44
Tmem106c
Trappc5
Uspl1
Vezt
Wdr95
Xab2
Zfp958
|
|
Liver weight, left
|
13.45 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Liver weight, right
|
15.14 |
12 |
6 |
5.6 |
0.98 |
7.81e-21 |
Cacul1
Cd99
Dennd10
Fam204a
Grk5
LOC103694460
Nanos1
Plekhs1
Prlhr
Rab11fip2
Sfxn4
Vti1a
|
|
Parametrial fat weight
|
16.19 |
55 |
31 |
28.7 |
0.99 |
0.00e+00 |
Ablim1
Acsl5
Afap1l2
Atrnl1
Bfsp1
Cacul1
Casp7
Ccdc186
Ces2c
Dclre1a
Dennd10
Dstn
Dtd1
Dzank1
Emx2os
Eno4
Fam204a
Fhip2a
Gfra1
Gpam
Grk5
Gucy2g
Habp2
Hspa12a
Kif16b
LOC102547573
LOC102550304
LOC102550729
LOC102551125
LOC120100065
LOC120100068
LOC120101717
LOC120101719
Nanos1
Nhlrc2
Nrap
Otor
Pcsk2
Plekhs1
Pnlip
Polr3f
Prdx3
Prlhr
Rab11fip2
Rbbp9
Rrbp1
Sec23b
Sfxn4
Smim26
Snrpb2
Tcf7l2
Trub1
Zdhhc6
Zfp133
Zfp950
|
|
Retroperitoneal fat weight
|
17.46 |
85 |
70 |
64.8 |
1 |
0.00e+00 |
Ablim1
Acsl5
Add3
Adrb1
Afap1l2
Apoh
Arsg
Atrnl1
Axin2
Bfsp1
Cacul1
Casp7
Ccdc186
Cd99
Cep112
Ces2c
Dclre1a
Dennd10
Dstn
Dtd1
Dzank1
Emx2os
Eno4
Esf1
Fam204a
Fhip2a
Flrt3
Gfra1
Gpam
Grk5
Gucy2g
Habp2
Hspa12a
Ism1
Kat14
Kif16b
Klhdc8b
LOC102547573
LOC102548513
LOC102550112
LOC102550304
LOC102550729
LOC102551125
LOC103694460
LOC120095201
LOC120100064
LOC120100065
LOC120100068
LOC120101717
LOC120101719
Macrod2
Mxi1
Nanos1
Ndufaf5
Nhlrc2
Nrap
Otor
Pcsk2
Pdcd4
Plekhs1
Pnlip
Polr3f
Prdx3
Prkca
Prlhr
Rab11fip2
Rbbp9
Rgs9
Rrbp1
Sec23b
Sfxn4
Shoc2
Smim26
Snrpb2
Sptbn5
Sptlc3
Tasp1
Tcf7l2
Tectb
Tex10
Trub1
Vti1a
Zdhhc6
Zfp133
Zfp950
|
|
Intraocular pressure
|
19.47 |
2 |
1 |
0.9 |
0 |
1.00e+00 |
LOC102548155
LOC103694460
|
|
Extensor digitorum longus weight
|
6.19 |
126 |
47 |
43.5 |
0.98 |
0.00e+00 |
Adcy6
Ankrd50
Anxa5
Arf3
Arhgef18
Arl5c
Asb8
Atp11b
Bbs7
Brca2
C7h12orf54
Cacnb1
Cacnb3
Camsap3
Ccdc65
Ccnt1
Cd209a
Cdk12
Cers4
Cfap206
Cisd3
Col2a1
Ctxn1
Cwc25
Dcun1d1
Ddx23
Dnajc22
Elavl1
Endou
Epop
Fat4
Fbxl20
Fermt3
Fkbp11
Fry
Grb7
Hdac7
Igfbp4
Insr
Jade1
Kansl2
Kcnh3
Kl
Kmt2d
Lamtor3
Larp1b
Lasp1
Lmbr1l
LOC102547266
LOC102548155
LOC102548389
LOC102549089
LOC102549494
LOC102552452
LOC102556302
LOC102557269
LOC103692978
LOC103694460
LOC108351548
LOC108352129
LOC120093728
LOC120093732
LOC120093830
LOC120095826
LOC120095871
LOC120095873
Lrrc8e
Map2k7
Mccc1
Mcemp1
Mcoln1
Med1
Med30
Mllt6
Msl1
N4bp2l1
N4bp2l2
Neurod2
Olr1877
Or8s2
Orc3
Ormdl3
Pced1b
Pcgf2
Pcp2
Pet100
Pex11g
Pfkm
Ppp1r1b
Prkag1
Qrfpr
Rapgef3
Rapgefl1
Rars2
RGD1359108
Rhebl1
Rnd1
Rpap3
Rpl19
Rpl23
Rps6ka4
Rxfp2
Senp1
Slc35a1
Slc48a1
Smarce1
Snapc2
Spmip11
Srcin1
Stac2
Stard13
Stard3
Stxbp2
Timm44
Tmem106c
Trappc5
Tuba1a
Tuba1b
Tuba1c
Vdr
Wipf2
Xab2
Zfp267
Zfp641
Zfp958
Zpbp2
|
|
Soleus weight
|
7.07 |
9 |
0 |
0 |
0.99 |
4.41e-13 |
Ablim1
Atrnl1
Gpam
Grk5
Klhdc8b
LOC120100068
Tectb
Trub1
Vti1a
|
|
Tibialis anterior weight
|
7.55 |
65 |
40 |
37 |
0.98 |
0.00e+00 |
Ankrd50
Anxa5
Arg2
Arhgef18
Atp11b
Atp6v1d
Bbs7
Brca2
Camsap3
Cd209a
Cers4
Cfap206
Cnr1
Ctxn1
Dcun1d1
Elavl1
Fat4
Fermt3
Fry
Insr
Jade1
Kl
Larp1b
LOC102547266
LOC102548389
LOC102549089
LOC102549494
LOC102552452
LOC102557269
LOC103690858
LOC103694460
LOC120095826
LOC120095871
LOC120095873
LOC120102853
Lrrc8e
Map2k7
Mccc1
Mcemp1
Mcoln1
Med30
N4bp2l1
N4bp2l2
Orc3
Pals1
Pcp2
Pet100
Pex11g
Pigh
Pnpla6
Qrfpr
Rars2
Retn
RGD1359108
Rps6ka4
Rxfp2
Slc35a1
Snapc2
Stard13
Stxbp2
Timm44
Trappc5
Xab2
Zfp267
Zfp958
|
|
Tibia length
|
8.03 |
123 |
21 |
19.4 |
0.99 |
0.00e+00 |
Adcy6
Als2cl
Amdhd1
Arf3
Arih2
Asb8
Atrip
C7h12orf54
Cacnb3
Camp
Ccdc12
Ccdc38
Ccdc51
Ccdc65
Ccnt1
Cdc25a
Cep83
Cep83os
Col2a1
Cspg5
Dclk3
Ddx23
Dhh
Dnajc22
Elk3
Elp6
Epm2aip1
Esf1
Fgd6
Fkbp11
Flrt3
Insr
Kansl2
Kif16b
Kif9
Klhdc8b
Klhl18
Kmt2d
Lmbr1l
LOC102546778
LOC102548155
LOC102548513
LOC102548848
LOC102548950
LOC102552504
LOC102553539
LOC102556302
LOC102556485
LOC103692978
LOC108351781
LOC120093556
LOC120093559
LOC120093563
LOC120093732
LOC120093830
LOC120095870
LOC120101717
Lrrc2
Lrrfip2
Lta4h
Macrod2
Map2k7
Map4
Mcoln1
Mir331
Mlh1
Myl3
N4bp2l1
N4bp2l2
Nbeal2
Nckipsd
Ndufa12
Ndufaf5
Ngp
Nme6
Nr2c1
Nradd
Ntn4
Olr1877
Or8s2
Otor
Pcp2
Pet100
Pfkfb4
Pfkm
Plxnb1
Plxnc1
Prkag1
Prph
Prss50
Pth1r
Ptpn23
Rapgef3
Rhebl1
Rnd1
Rpap3
Rtp3
Scap
Senp1
Setd2
Shisa5
Slc48a1
Smarcc1
Snapc2
Snrpb2
Snrpf
Socs2
Spats2
Spmip11
Sptlc3
Stxbp2
Tasp1
Tmcc3
Tmem106c
Trank1
Trex1
Tuba1a
Tuba1b
Tuba1c
Ube2n
Vezt
Wnt1
Zfp641
|
|
Number of licking bursts
|
24.53 |
1 |
1 |
0.9 |
0 |
1.00e+00 |
Klhdc8b
|
|
Patch foraging indifference point 0 sec
|
12.96 |
1 |
0 |
0 |
0 |
1.00e+00 |
LOC103694460
|
|
Indifference point function ln k
|
36.38 |
1 |
1 |
0.9 |
0 |
1.00e+00 |
Klhdc8b
|
|
Indifference point function log k
|
36.38 |
1 |
1 |
0.9 |
0 |
1.00e+00 |
Klhdc8b
|
|
Patch foraging total patch changes 0 sec
|
12.64 |
1 |
0 |
0 |
0 |
1.00e+00 |
LOC103694460
|
|
Locomotor activity
|
57.78 |
1 |
1 |
0.9 |
0 |
1.00e+00 |
Klhdc8b
|
|
Locomotor testing distance
|
64.52 |
1 |
1 |
0.9 |
0 |
1.00e+00 |
Klhdc8b
|
|
Reaction time number correct
|
18.64 |
1 |
1 |
0.9 |
0 |
1.00e+00 |
Klhdc8b
|
|
Reaction time trials correct on left
|
18.64 |
1 |
1 |
0.9 |
0 |
1.00e+00 |
Klhdc8b
|
|
Reaction time trials on left
|
19.51 |
1 |
1 |
0.9 |
0 |
1.00e+00 |
Klhdc8b
|
|
Reaction time omissions
|
23.75 |
1 |
1 |
0.9 |
0 |
1.00e+00 |
Klhdc8b
|
|
Reaction time false alarm rate
|
20.24 |
1 |
1 |
0.9 |
0 |
1.00e+00 |
Klhdc8b
|
|
Std. dev. reaction times
|
14.2 |
1 |
0 |
0 |
0 |
1.00e+00 |
Jade1
|
|
Reaction time trials completed
|
19.51 |
1 |
1 |
0.9 |
0 |
1.00e+00 |
Klhdc8b
|
|
Reaction time trials AUC
|
20.65 |
1 |
1 |
0.9 |
0 |
1.00e+00 |
Klhdc8b
|
|
Conditioned locomotion
|
28.4 |
1 |
1 |
0.9 |
0 |
1.00e+00 |
Klhdc8b
|
|
Cocaine response before conditioning
|
38.66 |
1 |
1 |
0.9 |
0 |
1.00e+00 |
Klhdc8b
|
|
Condit. Reinf. active responses
|
14.63 |
2 |
0 |
0 |
0 |
1.00e+00 |
Fam13b
Reep5
|
|
Condit. Reinf. lever reinforcers received
|
13.94 |
2 |
0 |
0 |
0 |
1.00e+00 |
Fam13b
Reep5
|
|
Pavlov. Cond. index score
|
15.43 |
1 |
0 |
0 |
0 |
1.00e+00 |
LOC103694460
|
|
Pavlov. Cond. magazine entry number
|
25.13 |
2 |
1 |
0.9 |
0 |
1.00e+00 |
Klhdc8b
LOC103694460
|
|
Conditioned reinforcement - inactives
|
16.77 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Intermitt. access day 15 inactive lever presses
|
30.78 |
1 |
1 |
0.9 |
0 |
1.00e+00 |
Klhdc8b
|
|
Intermittent access terminal intake (last 3 days)
|
24.03 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Short access day 10 total inactive lever presses
|
101.65 |
1 |
1 |
0.9 |
0 |
1.00e+00 |
Klhdc8b
|
|
Locomotion distance, session 1
|
23.98 |
1 |
1 |
0.9 |
0 |
1.00e+00 |
Klhdc8b
|
|
Locomotion distance, session 2
|
33.15 |
1 |
1 |
0.9 |
0 |
1.00e+00 |
Klhdc8b
|
|
Locomotion velocity, session 3
|
17.14 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Locomotion distance, session 8
|
52.78 |
1 |
1 |
0.9 |
0 |
1.00e+00 |
Klhdc8b
|
|
Degree of sensitization distance
|
24.75 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Condit. Reinf. active minus inactive responses
|
35.59 |
1 |
1 |
0.9 |
0 |
1.00e+00 |
Klhdc8b
|
|
Condit. Reinf. active-inactive response ratio
|
25.6 |
1 |
1 |
0.9 |
0 |
1.00e+00 |
Klhdc8b
|
|
Condit. Reinf. active responses
|
18.23 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Incentive salience index mean
|
26.23 |
1 |
1 |
0.9 |
0 |
1.00e+00 |
Klhdc8b
|
|
Time in familiar zone, hab. session 1
|
63.49 |
1 |
1 |
0.9 |
0 |
1.00e+00 |
Klhdc8b
|
|
Total zone transitions, hab. session 1
|
37.38 |
1 |
1 |
0.9 |
0 |
1.00e+00 |
Klhdc8b
|
|
Time in familiar zone, hab. session 2
|
27.1 |
1 |
1 |
0.9 |
0 |
1.00e+00 |
Klhdc8b
|
|
Total zone transitions, hab. session 2
|
69.58 |
1 |
1 |
0.9 |
0 |
1.00e+00 |
Klhdc8b
|
|
Total locomotion distance, hab. session 2
|
19.2 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Locomotion velocity, hab. session 2
|
21.15 |
1 |
1 |
0.9 |
0 |
1.00e+00 |
Klhdc8b
|
|
Novel to familiar place preference ratio
|
22.33 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Novelty place preference
|
20.99 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Total zone transitions, NPP test
|
59.86 |
2 |
1 |
0.9 |
0 |
1.00e+00 |
Klhdc8b
Trpc1
|
|
Total locomotion distance, NPP test
|
47.64 |
1 |
1 |
0.9 |
0 |
1.00e+00 |
Klhdc8b
|
|
Locomotion velocity, NPP test
|
51.12 |
1 |
1 |
0.9 |
0 |
1.00e+00 |
Klhdc8b
|
|
Pavlov. Cond. change in total contacts
|
69.48 |
1 |
1 |
0.9 |
0 |
1.00e+00 |
Klhdc8b
|
|
Pavlov. Cond. magazine entry number
|
15.14 |
1 |
0 |
0 |
0 |
1.00e+00 |
Fbxl20
|
|
Bone: apparent density
|
12.38 |
17 |
0 |
0 |
0.99 |
3.78e-52 |
Adcy6
Ddx23
Fry
Hmgb1
Hsph1
Kansl2
LOC102546827
LOC102548389
LOC102549494
LOC102553253
LOC120095871
LOC120095873
Mcemp1
Rnd1
Rxfp2
Trappc5
Zfp958
|
|
Bone surface
|
11.81 |
55 |
0 |
0 |
0.99 |
1.76e-227 |
Adcy6
Arf3
Asb8
C7h12orf54
Cacnb3
Ccdc65
Ccnt1
Col2a1
Ddx23
Dhh
Dnajc22
Faim2
Fkbp11
Fry
Hdac7
Hsph1
Kansl2
Kcnh3
Kmt2d
Lmbr1l
LOC100362820
LOC102546778
LOC102546827
LOC102548155
LOC102548389
LOC102549494
LOC102556302
LOC103692978
LOC108351548
LOC120093732
LOC120093830
LOC120095871
LOC120095873
Olr1877
Or8s2
Pfkm
Prkag1
Prph
Rapgef3
Rhebl1
Rnd1
Rxfp2
Senp1
Slc48a1
Snapc2
Spats2
Spmip11
Tmem106c
Trappc5
Tuba1a
Tuba1b
Tuba1c
Wnt1
Zfp641
Zfp958
|
|
Bone volume
|
12.31 |
11 |
0 |
0 |
1 |
2.90e-42 |
Fry
Hsph1
Lnc001
LOC102546827
LOC102548389
LOC102549494
LOC120095871
LOC120095873
Mcemp1
Rxfp2
Zfp958
|
|
Bone: connectivity density
|
13.37 |
12 |
1 |
0.9 |
1 |
4.35e-43 |
Fry
Hsph1
LOC102546827
LOC102548389
LOC102549494
LOC120095871
LOC120095873
Mcemp1
Rxfp2
Snapc2
Trappc5
Zfp958
|
|
Bone: cortical area
|
13.91 |
23 |
0 |
0 |
0.98 |
5.88e-50 |
Ablim1
Acsl5
Afap1l2
Cacul1
Casp7
Ces2c
Dclre1a
Dennd10
Emx2os
Fam204a
Fhip2a
Grk5
Habp2
LOC102547573
LOC102550729
LOC120100065
LOC120100068
Nhlrc2
Plekhs1
Prlhr
Rab11fip2
Sfxn4
Zfp950
|
|
Bone: cortical thickness
|
16.47 |
10 |
5 |
4.6 |
0.94 |
5.17e-14 |
Emx2os
Esf1
Ism1
LOC120100068
Macrod2
Ndufaf5
Prlhr
Rab11fip2
Sptlc3
Tasp1
|
|
Bone: endosteal estimation
|
8.88 |
2 |
0 |
0 |
0 |
1.00e+00 |
LOC102546778
Slc26a8
|
|
Bone: endosteal perimeter
|
18.12 |
2 |
1 |
0.9 |
0 |
1.00e+00 |
Cd99
LOC103694460
|
|
Bone: final force
|
12.04 |
5 |
0 |
0 |
0.93 |
2.19e-02 |
Faim2
LOC100362820
LOC102546778
Spats2
Zfp950
|
|
Bone: final moment
|
12.41 |
13 |
0 |
0 |
0.92 |
5.81e-08 |
Adcy6
Cacul1
Ddx23
Faim2
Grk5
Kansl2
Kcnh3
LOC100362820
LOC102546778
Prlhr
Sfxn4
Spats2
Zfp950
|
|
Bone: marrow area
|
8.76 |
2 |
0 |
0 |
0 |
1.00e+00 |
LOC102546778
Slc26a8
|
|
Bone: maximum diameter
|
11.7 |
14 |
1 |
0.9 |
0.99 |
2.80e-26 |
Ablim1
Acsl5
Afap1l2
Casp7
Ccdc186
Cd99
Dclre1a
Fhip2a
Habp2
LOC102547573
LOC103694460
LOC120100065
Nhlrc2
Plekhs1
|
|
Bone: maximum force
|
13.37 |
51 |
5 |
4.6 |
0.99 |
0.00e+00 |
Adcy6
Arf3
Asb8
C7h12orf54
Cacnb3
Cacul1
Ccdc65
Ccnt1
Col2a1
Ddx23
Dhh
Dnajc22
Endou
Faim2
Fkbp11
Hdac7
Kansl2
Kcnh3
Kmt2d
Lmbr1l
LOC100362820
LOC102546778
LOC102548155
LOC102556302
LOC103692978
LOC108351548
LOC120093728
LOC120093732
LOC120093830
Olr1877
Or8s2
Pced1b
Pfkm
Prkag1
Prlhr
Prph
Rapgef3
Rhebl1
Rnd1
Rpap3
Senp1
Slc48a1
Spats2
Spmip11
Tmem106c
Tuba1a
Tuba1b
Tuba1c
Vdr
Wnt1
Zfp641
|
|
Bone: maximum moment
|
13.37 |
54 |
7 |
6.5 |
0.98 |
2.27e-278 |
Adcy6
Arf3
Asb8
C7h12orf54
Cacnb3
Cacul1
Ccdc65
Ccnt1
Col2a1
Ddx23
Dhh
Dnajc22
Endou
Faim2
Fkbp11
Grk5
Hdac7
Kansl2
Kcnh3
Kmt2d
Lmbr1l
LOC100362820
LOC102546778
LOC102548155
LOC102556302
LOC103692978
LOC108351548
LOC120093728
LOC120093732
LOC120093830
Olr1877
Or8s2
Pced1b
Pfkm
Prkag1
Prlhr
Prph
Rapgef3
Rhebl1
Rnd1
Rpap3
Senp1
Sfxn4
Slc48a1
Spats2
Spmip11
Tmem106c
Tuba1a
Tuba1b
Tuba1c
Vdr
Wnt1
Zfp641
Zfp950
|
|
Bone: minimum diameter
|
10.22 |
1 |
0 |
0 |
0 |
1.00e+00 |
LOC102546778
|
|
Bone: periosteal estimation
|
12.03 |
59 |
7 |
6.5 |
0.97 |
1.04e-239 |
Acsl5
Adcy6
Arf3
Asb8
C7h12orf54
Cacnb3
Cacul1
Ccdc65
Ccnt1
Col2a1
Ddx23
Dennd10
Dhh
Dnajc22
Endou
Faim2
Fam204a
Fkbp11
Grk5
Hdac7
Kansl2
Kcnh3
Kmt2d
Lmbr1l
LOC100362820
LOC102546778
LOC102548155
LOC102556302
LOC103692978
LOC108351548
LOC120093728
LOC120093732
LOC120093830
Olr1877
Or8s2
Pced1b
Pfkm
Plekhs1
Prkag1
Prlhr
Prph
Rab11fip2
Rapgef3
Rhebl1
Rnd1
Rpap3
Senp1
Sfxn4
Slc48a1
Spats2
Spmip11
Tmem106c
Tuba1a
Tuba1b
Tuba1c
Vdr
Wnt1
Zfp641
Zfp950
|
|
Bone: periosteal perimeter
|
12.91 |
43 |
3 |
2.8 |
0.96 |
1.07e-107 |
Ablim1
Acsl5
Afap1l2
Asb8
Atrnl1
Cacnb3
Cacul1
Casp7
Ccdc186
Ces2c
Dclre1a
Dennd10
Fam204a
Fhip2a
Gfra1
Grk5
Habp2
Hdac7
Kansl2
Lmbr1l
LOC102547573
LOC102548155
LOC102550729
LOC102551125
LOC103694460
LOC120093830
LOC120100065
Nhlrc2
Nrap
Olr1877
Pfkm
Plekhs1
Rapgef3
Rnd1
Rpap3
Senp1
Sfxn4
Slc48a1
Tmem106c
Trub1
Tuba1c
Zfp641
Zfp950
|
|
Bone: stiffness
|
16.25 |
50 |
32 |
29.6 |
0.99 |
0.00e+00 |
Adcy6
Arf3
Asb8
C7h12orf54
Cacnb3
Ccdc65
Ccnt1
Col2a1
Ddx23
Dhh
Dnajc22
Endou
Faim2
Fkbp11
Hdac7
Kansl2
Kcnh3
Klhdc8b
Kmt2d
Lmbr1l
LOC100362820
LOC102546778
LOC102548155
LOC102556302
LOC103692978
LOC108351548
LOC120093728
LOC120093732
LOC120093830
Olr1877
Or8s2
Pced1b
Pfkm
Prkag1
Prph
Rapgef3
Rhebl1
Rnd1
Rpap3
Senp1
Slc48a1
Spats2
Spmip11
Tmem106c
Tuba1a
Tuba1b
Tuba1c
Vdr
Wnt1
Zfp641
|
|
Bone: trabecular number
|
13.03 |
4 |
0 |
0 |
1 |
5.26e-10 |
Hsph1
LOC102546827
LOC102549494
Rxfp2
|
|
Bone: trabecular thickness
|
16.33 |
3 |
0 |
0 |
1 |
1.03e-06 |
Cacul1
Prlhr
Sfxn4
|
|
Time to tail flick, vehicle, after self-admin
|
37.57 |
1 |
1 |
0.9 |
0 |
1.00e+00 |
Klhdc8b
|
|
Total heroin consumption
|
18.89 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Delay disc. indifference point, 0s delay
|
28.7 |
1 |
1 |
0.9 |
0 |
1.00e+00 |
Klhdc8b
|
|
Delay disc. indifference point, 4s delay
|
41.82 |
1 |
1 |
0.9 |
0 |
1.00e+00 |
Klhdc8b
|
|
Rest time, locomotor task time 1
|
22.55 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Distance moved, locomotor task time 1
|
31.01 |
1 |
1 |
0.9 |
0 |
1.00e+00 |
Klhdc8b
|
|
Vertical activity count, locomotor time 1
|
49.01 |
1 |
1 |
0.9 |
0 |
1.00e+00 |
Klhdc8b
|
|
Bouts of movement, locomotor time 2
|
19.85 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Total resting periods, locomotor time 2
|
20.22 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Rest time, locomotor task time 2
|
25.34 |
1 |
1 |
0.9 |
0 |
1.00e+00 |
Klhdc8b
|
|
Distance moved, locomotor task time 2
|
42.39 |
1 |
1 |
0.9 |
0 |
1.00e+00 |
Klhdc8b
|
|
Vertical activity count, locomotor time 2
|
54.92 |
1 |
1 |
0.9 |
0 |
1.00e+00 |
Klhdc8b
|
|
Weight adjusted by age
|
30.32 |
1 |
1 |
0.9 |
0 |
1.00e+00 |
Klhdc8b
|
|
Food seeking constrained by brief footshock
|
21.42 |
2 |
2 |
1.9 |
1 |
6.39e-30 |
LOC102557269
RGD1359108
|
|
Cd content in liver
|
20.17 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Cu content in liver
|
31.36 |
2 |
1 |
0.9 |
0 |
1.00e+00 |
Cd99
LOC103694460
|
|
Fe content in liver
|
33.68 |
2 |
2 |
1.9 |
0 |
1.00e+00 |
Klhdc8b
LOC103694460
|
|
K content in liver
|
34.63 |
2 |
1 |
0.9 |
0 |
1.00e+00 |
Cd99
LOC103694460
|
|
Rb content in liver
|
54.46 |
3 |
3 |
2.8 |
0 |
1.00e+00 |
Cd99
Klhdc8b
LOC103694460
|
|
Sr content in liver
|
22.32 |
2 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
LOC103694460
|