Liver weight, left
Tags: Physiology · Liver · Weight
Project: dissection
1 locus · 1 gene with independent associations · 1 total associated gene
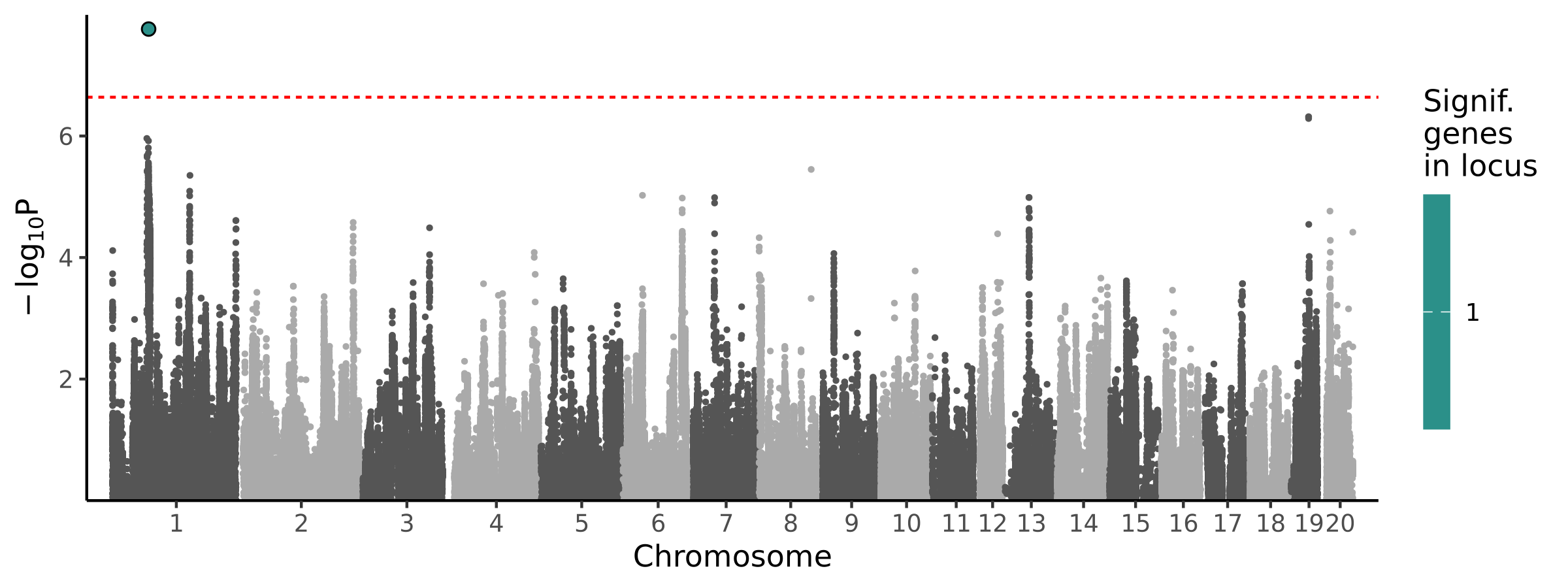
Significant Loci
| # | Chr | Start pos | End pos | # assoc genes | # joint models | Best TWAS P | Best GWAS P | Cond GWAS P | Joint genes |
|---|---|---|---|---|---|---|---|---|---|
| 1 | chr1 | 76658582 | 78051660 | 1 | 1 | 1.74e-08 | 2.87e-06 | 1.13e-75 | LOC120099769 |
Pleiotropic Associations
Associations by panel
| Tissue | RNA modality | # hits | % hits/tests | Avg chisq |
|---|---|---|---|---|
| Adipose | alternative polyA | 0 | 0 | 1.67 |
| Adipose | alternative TSS | 0 | 0 | 1.54 |
| Adipose | gene expression | 0 | 0 | 1.61 |
| Adipose | isoform ratio | 0 | 0 | 1.56 |
| Adipose | intron excision ratio | 0 | 0 | 1.51 |
| Adipose | mRNA stability | 0 | 0 | 1.56 |
| BLA | alternative polyA | 0 | 0 | 1.55 |
| BLA | alternative TSS | 0 | 0 | 1.67 |
| BLA | gene expression | 0 | 0 | 1.62 |
| BLA | isoform ratio | 0 | 0 | 1.68 |
| BLA | intron excision ratio | 0 | 0 | 1.57 |
| BLA | mRNA stability | 0 | 0 | 1.66 |
| Brain | alternative polyA | 0 | 0 | 1.57 |
| Brain | alternative TSS | 0 | 0 | 1.64 |
| Brain | gene expression | 1 | 0 | 1.58 |
| Brain | isoform ratio | 0 | 0 | 1.62 |
| Brain | intron excision ratio | 0 | 0 | 1.57 |
| Brain | mRNA stability | 0 | 0 | 1.57 |
| Eye | alternative polyA | 0 | 0 | 1.7 |
| Eye | alternative TSS | 0 | 0 | 1.78 |
| Eye | gene expression | 0 | 0 | 1.69 |
| Eye | isoform ratio | 0 | 0 | 1.76 |
| Eye | intron excision ratio | 0 | 0 | 1.55 |
| Eye | mRNA stability | 0 | 0 | 1.57 |
| IL | alternative polyA | 0 | 0 | 1.55 |
| IL | alternative TSS | 0 | 0 | 1.83 |
| IL | gene expression | 0 | 0 | 1.6 |
| IL | isoform ratio | 0 | 0 | 1.77 |
| IL | intron excision ratio | 0 | 0 | 1.59 |
| IL | mRNA stability | 0 | 0 | 1.65 |
| LHb | alternative polyA | 0 | 0 | 1.61 |
| LHb | alternative TSS | 0 | 0 | 1.61 |
| LHb | gene expression | 0 | 0 | 1.67 |
| LHb | isoform ratio | 0 | 0 | 1.64 |
| LHb | intron excision ratio | 0 | 0 | 1.54 |
| LHb | mRNA stability | 0 | 0 | 1.67 |
| Liver | alternative polyA | 0 | 0 | 1.63 |
| Liver | alternative TSS | 0 | 0 | 1.61 |
| Liver | gene expression | 0 | 0 | 1.6 |
| Liver | isoform ratio | 0 | 0 | 1.56 |
| Liver | intron excision ratio | 0 | 0 | 1.49 |
| Liver | mRNA stability | 0 | 0 | 1.64 |
| NAcc | alternative polyA | 0 | 0 | 1.55 |
| NAcc | alternative TSS | 0 | 0 | 1.64 |
| NAcc | gene expression | 0 | 0 | 1.62 |
| NAcc | isoform ratio | 0 | 0 | 1.61 |
| NAcc | intron excision ratio | 0 | 0 | 1.6 |
| NAcc | mRNA stability | 0 | 0 | 1.59 |
| OFC | alternative polyA | 0 | 0 | 1.64 |
| OFC | alternative TSS | 0 | 0 | 1.83 |
| OFC | gene expression | 0 | 0 | 1.68 |
| OFC | isoform ratio | 0 | 0 | 1.73 |
| OFC | intron excision ratio | 0 | 0 | 1.49 |
| OFC | mRNA stability | 0 | 0 | 1.7 |
| PL | alternative polyA | 0 | 0 | 1.59 |
| PL | alternative TSS | 0 | 0 | 1.68 |
| PL | gene expression | 0 | 0 | 1.61 |
| PL | isoform ratio | 0 | 0 | 1.63 |
| PL | intron excision ratio | 0 | 0 | 1.55 |
| PL | mRNA stability | 0 | 0 | 1.61 |
| pVTA | alternative polyA | 0 | 0 | 1.53 |
| pVTA | alternative TSS | 0 | 0 | 1.64 |
| pVTA | gene expression | 0 | 0 | 1.6 |
| pVTA | isoform ratio | 0 | 0 | 1.6 |
| pVTA | intron excision ratio | 0 | 0 | 1.53 |
| pVTA | mRNA stability | 0 | 0 | 1.54 |
| RMTg | alternative polyA | 0 | 0 | 1.55 |
| RMTg | alternative TSS | 0 | 0 | 1.67 |
| RMTg | gene expression | 0 | 0 | 1.62 |
| RMTg | isoform ratio | 0 | 0 | 1.44 |
| RMTg | intron excision ratio | 0 | 0 | 1.54 |
| RMTg | mRNA stability | 0 | 0 | 1.42 |