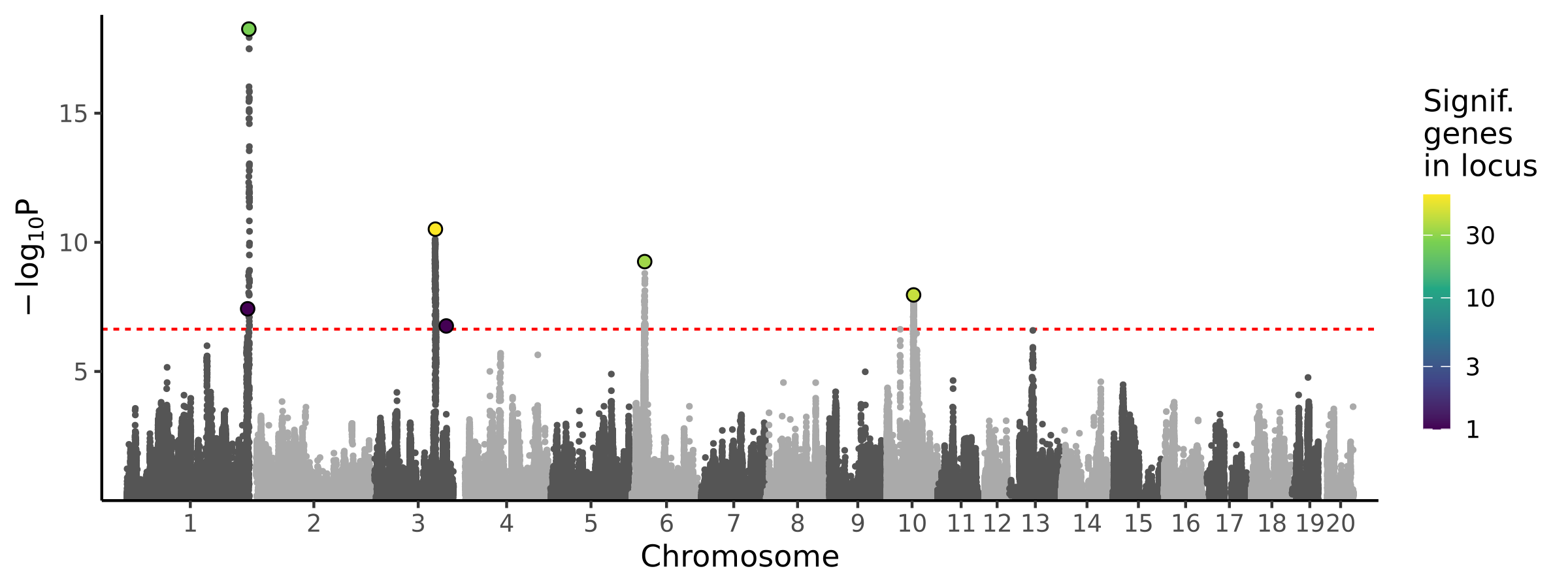
|
BMI with tail
|
19.84 |
74 |
70 |
291.7 |
0.64 |
8.19e-64 |
Ablim1
Acsl5
Afap1l2
Atrnl1
Banf2
Cacul1
Casp7
Ccdc186
Ces2c
Dclre1a
Dennd10
Dstn
Dtd1
Dzank1
Emx2os
Eno4
Fam204a
Fhip2a
Gfra1
Gpam
Grk5
Gucy2g
Habp2
Hspa12a
Kat14
Kif16b
LOC102547573
LOC102550304
LOC102551125
LOC102553657
LOC102556644
LOC103691380
LOC120097692
LOC120100061
LOC120100065
LOC120100068
LOC120101717
LOC120101719
LOC134485255
LOC134485257
LOC134485258
LOC134485266
LOC134485267
LOC134485268
LOC134486165
LOC134486454
LOC134486455
LOC134486456
Nanos1
Nhlrc2
Nrap
Otor
Pcsk2
Plekhs1
Pnlip
Polr3f
Prdx3
Prlhr
Rab11fip2
Rbbp9
Retreg2
Rrbp1
Scp2d1
Sec23b
Sfxn4
Smim26
Snrpb2
Tcf7l2
Tectb
Trub1
Vti1a
Zdhhc6
Zfp133
Zfp933l1
|
|
BMI without tail
|
301.68 |
68 |
41 |
170.8 |
0.37 |
1.59e-17 |
Ablim1
Acsl5
Afap1l2
Atrnl1
Banf2
Cacul1
Casp7
Ccdc186
Ces2c
Dclre1a
Dennd10
Dstn
Dtd1
Dzank1
Emx2os
Eno4
Fam204a
Fhip2a
Gfra1
Gpam
Grk5
Gucy2g
Habp2
Hspa12a
Kat14
Kif16b
LOC102547573
LOC102550304
LOC102551125
LOC120097692
LOC120100061
LOC120100065
LOC120100068
LOC120101717
LOC120101719
LOC134485255
LOC134485257
LOC134485258
LOC134485266
LOC134485267
LOC134486165
LOC134486454
LOC134486455
LOC134486456
Nanos1
Nhlrc2
Nrap
Otor
Pcsk2
Plekhs1
Polr3f
Prdx3
Prlhr
Rab11fip2
Rbbp9
Retreg2
Rrbp1
Scp2d1
Sec23b
Sfxn4
Smim26
Snrpb2
Tcf7l2
Trub1
Vti1a
Zdhhc6
Zfp133
Zfp933l1
|
|
Body weight
|
13.17 |
74 |
72 |
300 |
0.97 |
0.00e+00 |
Ablim1
Acsl5
Afap1l2
Atrnl1
Banf2
Cacul1
Casp7
Ccdc186
Ces2c
Dclre1a
Dennd10
Dstn
Dtd1
Dzank1
Emx2os
Eno4
Fam204a
Fhip2a
Gfra1
Gpam
Grk5
Gucy2g
Habp2
Hspa12a
Kat14
Kif16b
LOC102547573
LOC102550304
LOC102551125
LOC102553657
LOC102556644
LOC103691380
LOC120097692
LOC120100061
LOC120100065
LOC120100068
LOC120101717
LOC120101719
LOC134485255
LOC134485257
LOC134485258
LOC134485266
LOC134485267
LOC134485268
LOC134486165
LOC134486454
LOC134486455
LOC134486456
Nanos1
Nhlrc2
Nrap
Otor
Pcsk2
Plekhs1
Pnlip
Polr3f
Prdx3
Prlhr
Rab11fip2
Rbbp9
Retreg2
Rrbp1
Scp2d1
Sec23b
Sfxn4
Smim26
Snrpb2
Tcf7l2
Tectb
Trub1
Vti1a
Zdhhc6
Zfp133
Zfp933l1
|
|
Epididymis fat weight
|
101.88 |
148 |
145 |
604.2 |
0.46 |
4.79e-63 |
4933427D14Rikl
Abhd1
Ablim1
Acsl5
Adcy3
Afap1l2
Agbl5
Asxl2
Atad2b
Atraid
Atrnl1
Banf2
Cacul1
Cad
Casp7
Ccdc186
Cenpo
Ces2c
Cgref1
Cimip2c
Clip4
Cyb5d2
Dclre1a
Dennd10
Dnajc27
Dnmt3a
Dpysl5
Drc1
Dstn
Dtd1
Dtnb
Dzank1
Efr3b
Eif2b4
Emx2os
Eno4
Fam204a
Fam228b
Fhip2a
Fkbp1b
Fndc4
Fosl2
Garem2
Gfra1
Gpam
Gpn1
Grk5
Gtf3c2
Gucy2g
Habp2
Hadha
Hadhb
Hspa12a
Ift172
Itgae
Itsn2
Kat14
Kcnk3
Khk
Kif16b
Kif3c
Klhl29
LOC102547573
LOC102548914
LOC102550304
LOC102551125
LOC102553657
LOC102556644
LOC103691380
LOC103692581
LOC120097692
LOC120100061
LOC120100065
LOC120100068
LOC120101717
LOC120101719
LOC120103488
LOC120103494
LOC120103495
LOC120103497
LOC120103502
LOC134479112
LOC134479333
LOC134479335
LOC134485255
LOC134485257
LOC134485258
LOC134485266
LOC134485267
LOC134485268
LOC134486165
LOC134486454
LOC134486455
LOC134486456
Mapre3
Mpv17
Nanos1
Ncoa1
Nhlrc2
Nrap
Nrbp1
Ost4
Otof
Otor
Pcare
Pcsk2
Pfn4
Plb1
Plekhs1
Pnlip
Polr3f
Ppp1cb
Prdx3
Preb
Prlhr
Rab10
Rab11fip2
Rbbp9
Rbks
Retreg2
Rrbp1
Scp2d1
Sec23b
Sf3b6
Sfxn4
Slc30a3
Slc35f6
Slc4a1ap
Slc5a6
Smim26
Smtnl2
Snrpb2
Snx17
Spdya
Tcf7l2
Tectb
Tekt1
Tmem214
Togaram2
Trim54
Trub1
Ubxn2a
Vti1a
Yipf4l1
Zdhhc6
Zfp133
Zfp513
Zfp933l1
|
|
Glucose
|
1188.51 |
2 |
1 |
4.2 |
0 |
1.00e+00 |
Ces2c
Retreg2
|
|
Heart weight
|
296.4 |
12 |
4 |
16.7 |
-0.31 |
3.39e-02 |
Cacul1
Ces2c
Dennd10
Grk5
LOC120100068
LOC134485267
Nanos1
Prlhr
Rab11fip2
Retreg2
Sfxn4
Zfp933l1
|
|
Left kidney weight
|
1940.31 |
18 |
14 |
58.3 |
-0.43 |
1.55e-04 |
Cacul1
Casp7
Ces2c
Dennd10
Emx2os
Gpam
Grk5
LOC120100068
LOC134485266
LOC134485267
Nanos1
Prdx3
Prlhr
Rab11fip2
Retreg2
Sfxn4
Vti1a
Zfp933l1
|
|
Right kidney weight
|
1129.48 |
27 |
15 |
62.5 |
-0.38 |
8.36e-05 |
Ablim1
Acsl5
Cacul1
Casp7
Ces2c
Dennd10
Emx2os
Gpam
Grk5
Habp2
LOC120100068
LOC134485255
LOC134485266
LOC134485267
Nanos1
Nhlrc2
Nrap
Plekhs1
Prdx3
Prlhr
Rab11fip2
Retreg2
Sfxn4
Tcf7l2
Vti1a
Zdhhc6
Zfp933l1
|
|
Tail length
|
92117.69 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Length with tail
|
174.8 |
8 |
1 |
4.2 |
0.77 |
5.00e-08 |
Kif16b
LOC120101719
Otor
Pcsk2
Retreg2
Rrbp1
Sec23b
Snrpb2
|
|
Length without tail
|
1473.44 |
27 |
2 |
8.3 |
-0.46 |
7.18e-06 |
Acsl5
Cacul1
Casp7
Dclre1a
Gpam
Grk5
Gucy2g
Habp2
Kif16b
LOC102553657
LOC120100061
LOC120101719
LOC134485255
LOC134486165
Nanos1
Nhlrc2
Nrap
Otor
Prlhr
Rab11fip2
Retreg2
Sfxn4
Snrpb2
Tcf7l2
Tectb
Vti1a
Zdhhc6
|
|
Liver weight, left
|
286.48 |
16 |
6 |
25 |
-0.32 |
2.13e-02 |
Cacul1
Casp7
Ces2c
Dennd10
Emx2os
Grk5
LOC120100068
LOC134485266
LOC134485267
Nanos1
Prdx3
Prlhr
Rab11fip2
Retreg2
Sfxn4
Zfp933l1
|
|
Liver weight, right
|
12.1 |
42 |
1 |
4.2 |
0.6 |
9.06e-15 |
Abhd1
Acsl5
Atraid
Cacul1
Cad
Casp7
Clip4
Dpysl5
Eif2b4
Fndc4
Fosl2
Gpam
Gpn1
Gtf3c2
Gucy2g
Ift172
Khk
LOC102548914
LOC120103488
LOC134485255
Mpv17
Nrbp1
Otof
Pcare
Plb1
Ppp1cb
Preb
Prlhr
Rab11fip2
Rbks
Retreg2
Sfxn4
Slc30a3
Slc4a1ap
Slc5a6
Snx17
Spdya
Tcf7l2
Togaram2
Trim54
Vti1a
Zdhhc6
|
|
Retroperitoneal fat weight
|
38.52 |
144 |
143 |
595.8 |
0.88 |
0.00e+00 |
Abhd1
Ablim1
Acsl5
Adcy3
Afap1l2
Agbl5
Asxl2
Atad2b
Atraid
Atrnl1
Banf2
Cacul1
Cad
Casp7
Ccdc186
Cenpo
Ces2c
Cgref1
Cgrrf1
Cimip2c
Clip4
Dclre1a
Dennd10
Dnajc27
Dnmt3a
Dpysl5
Drc1
Dstn
Dtd1
Dtnb
Dzank1
Efr3b
Eif2b4
Emx2os
Eno4
Fam204a
Fam228b
Fhip2a
Fkbp1b
Fndc4
Fosl2
Garem2
Gfra1
Gpam
Gpn1
Grk5
Gtf3c2
Gucy2g
Habp2
Hadha
Hadhb
Hspa12a
Ift172
Itsn2
Kat14
Kcnk3
Khk
Kif16b
Kif3c
Klhl29
LOC102547573
LOC102548914
LOC102550304
LOC102551125
LOC102553657
LOC102556644
LOC103691380
LOC103692581
LOC120097692
LOC120100061
LOC120100065
LOC120100068
LOC120101717
LOC120101719
LOC120103488
LOC120103494
LOC120103495
LOC120103497
LOC120103502
LOC134479112
LOC134479333
LOC134479335
LOC134485255
LOC134485257
LOC134485258
LOC134485266
LOC134485267
LOC134485268
LOC134486165
LOC134486454
LOC134486455
LOC134486456
Mapre3
Mpv17
Nanos1
Ncoa1
Nhlrc2
Nrap
Nrbp1
Ost4
Otof
Otor
Pcare
Pcsk2
Pfn4
Plb1
Plekhs1
Pnlip
Polr3f
Ppp1cb
Prdx3
Preb
Prlhr
Rab10
Rab11fip2
Rbbp9
Rbks
Retreg2
Rrbp1
Scp2d1
Sec23b
Sf3b6
Sfxn4
Slc30a3
Slc35f6
Slc4a1ap
Slc5a6
Smim26
Snrpb2
Snx17
Spdya
Tcf7l2
Tectb
Tmem214
Togaram2
Trim54
Trub1
Ubxn2a
Vti1a
Yipf4l1
Zdhhc6
Zfp133
Zfp513
Zfp933l1
|
|
Intraocular pressure
|
40071.91 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Extensor digitorum longus weight
|
35.86 |
35 |
1 |
4.2 |
0.17 |
3.80e-02 |
Abhd1
Agbl5
Atraid
Cad
Cgref1
Clip4
Dpysl5
Eif2b4
Fndc4
Fosl2
Gpn1
Gtf3c2
Itgae
Khk
LOC102548914
LOC120100068
LOC120103488
Mapre3
Mpv17
Nrbp1
Ost4
Otof
Pcare
Plb1
Ppp1cb
Preb
Rbks
Retreg2
Slc30a3
Slc35f6
Slc5a6
Snx17
Spdya
Togaram2
Trim54
|
|
Soleus weight
|
13.88 |
13 |
1 |
4.2 |
0.75 |
2.04e-03 |
Ablim1
Acsl5
Clip4
Eno4
Fosl2
Gucy2g
LOC134485258
LOC134485266
Pcare
Ppp1cb
Rab11fip2
Retreg2
Togaram2
|
|
Tibialis anterior weight
|
1631.05 |
3 |
1 |
4.2 |
0 |
1.00e+00 |
4933427D14Rikl
Itgae
Retreg2
|
|
Tibia length
|
224.56 |
5 |
1 |
4.2 |
0.93 |
4.56e-07 |
Kif16b
LOC134486165
Otor
Retreg2
Snrpb2
|
|
Number of licking bursts
|
157119.65 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Food consumed during 24 hour testing period
|
106196.53 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Times rat made contact with spout
|
305.36 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Mean time between licks in bursts
|
199507.71 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Mean num. licks in bursts
|
142600.29 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Std. dev. time between licks in bursts
|
94042.02 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Water consumed over 24 hours
|
103408.69 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Patch foraging indifference point 0 sec
|
115003.89 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Patch foraging indifference point AUC
|
133114.6 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Indifference point function ln k
|
142006.4 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Indifference point function log k
|
142006.4 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Patch foraging total patch changes 0 sec
|
60135.1 |
2 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
Sfxn4
|
|
Patch foraging total patch changes 12 sec
|
10848.8 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Patch foraging total patch changes 18 sec
|
117768.16 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Patch foraging total patch changes 24 sec
|
167090.22 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Patch foraging total patch changes 6 sec
|
16324.9 |
2 |
1 |
4.2 |
0 |
1.00e+00 |
LOC134485258
Retreg2
|
|
Patch foraging time to switch 0 sec
|
156618.53 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Patch foraging water rate 0 sec
|
148807.58 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Patch foraging water rate 12 sec
|
161858.08 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Patch foraging water rate 18 sec
|
6082.71 |
8 |
1 |
4.2 |
-0.83 |
3.33e-05 |
Dtd1
Dzank1
Kat14
Rbbp9
Retreg2
Scp2d1
Sec23b
Smim26
|
|
Patch foraging water rate 24 sec
|
4691.77 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Patch foraging water rate 6 sec
|
3954.86 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Locomotor activity
|
12.34 |
1 |
0 |
0 |
0 |
1.00e+00 |
Retreg2
|
|
Locomotor testing distance
|
89448.55 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Locomotor testing rearing
|
152414.81 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Light reinforcement 1
|
155242.72 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Light reinforcement 2
|
107899.08 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Reaction time number correct
|
153465.22 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Reaction time mean minus median
|
205810.62 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Reaction time mean minus median AUC
|
208396.98 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Reaction time num false alarms
|
57075.22 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Reaction time num false alarms AUC
|
106496.51 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Reaction time trials correct on left
|
153465.22 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Reaction time trials on left
|
149991.58 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Reaction time mean
|
94380.53 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Reaction time mean AUC
|
113341.21 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Median of all reaction times
|
31417.12 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Reaction time omissions
|
34267.91 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Reaction time false alarm rate
|
891.26 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Reaction time premature initiation rate
|
36877.02 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Reaction time premature initiations
|
138718.63 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Std. dev. reaction times
|
149487.71 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Reaction time trials completed
|
149991.58 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Reaction time trials AUC
|
151696.42 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Social response proportion
|
35302.65 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Social responses
|
5682.08 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Social time
|
44238.34 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Conditioned locomotion
|
6074.35 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Cocaine response after cond. corrected
|
65812.39 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Cocaine response after cond. not corrected
|
64247.35 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Cocaine response before conditioning
|
2800.41 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Saline control response
|
12962.78 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Condit. Reinf. active minus inactive responses
|
53410.48 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Condit. Reinf. active-inactive response ratio
|
91582.24 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Condit. Reinf. active responses
|
9691.24 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Condit. Reinf. inactive responses
|
42106.18 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Condit. Reinf. lever presses
|
15873.43 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Condit. Reinf. lever reinforcers received
|
2000.77 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Pavlov. Cond. lever latency
|
10912.53 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Pavlov. Cond. magazine entry latency
|
49418.79 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Pavlov. Cond. change in total contacts
|
114781.94 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Pavlov. Cond. index score
|
47988.08 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Pavlov. Cond. latency score
|
58476.16 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Pavlov. Cond. lever contacts
|
22069.78 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Pavlov. Cond. magazine entry number
|
16773.11 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Pavlov. Cond. intertrial magazine entries
|
104933.78 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Pavlov. Cond. lever-magazine prob. diff.
|
44871.67 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Pavlov. Cond. response bias
|
5169.79 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Conditioned reinforcement - actives
|
191492.54 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Conditioned reinforcement - inactives
|
155032.91 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Conditioned locomotion
|
41346.85 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Intermittent access intake day 1-15 change
|
210605.99 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Intermittent access intake escalation
|
209543.53 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Intermittent access intake escalation 2
|
107995.31 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Intermitt. access day 1 inactive lever presses
|
236542.72 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Intermitt. access day 15 inactive lever presses
|
208241.66 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Intermitt. access escalation Index
|
184996.65 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Intermittent access day 1 total infusions
|
155525.68 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Intermittent access day 15 total infusions
|
131485.34 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Intermittent access terminal intake (last 3 days)
|
83205.14 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Intermittent access total infusions
|
675.9 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Intermittent access day 1 locomotion
|
104459.41 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Intermittent access day 15 total locomotion
|
177836.92 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Intermittent access total locomotion
|
711.59 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Intermittent access standard deviation
|
201906.25 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Cocaine induced anxiety
|
160280.46 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Post-drug Anxiety
|
20460.83 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Baseline Anxiety
|
51741.5 |
2 |
1 |
4.2 |
0 |
1.00e+00 |
Gmppa
Retreg2
|
|
Lifetime Intake
|
6830.03 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Incentive Sensitization - Responses
|
163579.25 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Incentive Sensitization - Breakpoint
|
142790.8 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Progressive ratio test 1 active lever presses
|
99872.69 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Progressive ratio test 1 breakpoint
|
96247.61 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Progressive ratio test 1 inactive lever presses
|
151100.07 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Progressive ratio test 2 active lever presses
|
57222.6 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Progressive ratio test 2 breakpoint
|
34975.96 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Progressive ratio test 2 inactive lever presses
|
164104.71 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Total sessions with >9 infusions
|
7953.9 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Short access day 1 total inactive lever presses
|
114392.27 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Short access day 10 total inactive lever presses
|
217140.65 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Short access day 1 total infusions
|
111479.43 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Short access day 10 total infusions
|
76293.54 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Short access total infusions
|
33946.03 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Short access day 1 locomotion
|
84055.65 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Short access day 10 total locomotion
|
181597.84 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Short access total locomotion
|
68266.59 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Compulsive drug intake
|
65439.45 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
One hour access (0.3 mA shock)
|
9669.44 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
One hour access (shock baseline)
|
140263.4 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Number of responses in last shaping day
|
51289.19 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Context. condit. distance diff. score
|
970.59 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Locomotion velocity, session 1
|
14615.25 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Locomotion distance, session 1
|
4416.18 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Locomotion velocity, session 2
|
9358 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Locomotion distance, session 2
|
2393.66 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Locomotion velocity, session 3
|
2332.53 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Locomotion distance, session 3
|
16615.78 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Stereotopy head waving bouts, day 3
|
5866.4 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Stereotopy head waving duration, day 3
|
12131.92 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Locomotion velocity, session 7
|
1310.17 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Locomotion distance, session 7
|
3744.54 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Stereotopy head waving bouts, day 7
|
10466.92 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Stereotopy head waving duration, day 7
|
8847.36 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Locomotion velocity, session 8
|
1892.98 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Locomotion distance, session 8
|
673.32 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Degree of sensitization distance
|
15309.16 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Degree of sensitization stereotypy
|
8496.38 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Condit. Reinf. active minus inactive responses
|
4120.78 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Condit. Reinf. active-inactive response ratio
|
1097.66 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Condit. Reinf. active responses
|
11664.93 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Condit. Reinf. inactive responses
|
19375.3 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Incentive salience index mean
|
14781.83 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Condit. Reinf. lever presses
|
21433.81 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Time in familiar zone, hab. session 1
|
41873.03 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Time in novel zone, hab. session 1
|
35522.88 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Total zone transitions, hab. session 1
|
1818.53 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Total locomotion distance, hab. session 1
|
1687 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Locomotion velocity, hab. session 1
|
1131.06 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Time in familiar zone, hab. session 2
|
23379.25 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Time in novel zone, hab. session 2
|
24924.95 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Total zone transitions, hab. session 2
|
280.53 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Total locomotion distance, hab. session 2
|
96.8 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Locomotion velocity, hab. session 2
|
145.53 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Novel to familiar place preference ratio
|
55508.81 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Novelty place preference
|
54383.96 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Time in novel zone, NPP test
|
40844.77 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Total zone transitions, NPP test
|
8230.03 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Total locomotion distance, NPP test
|
7114 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Locomotion velocity, NPP test
|
4041.22 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Pavlov. Cond. lever latency
|
2195.61 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Pavlov. Cond. change in total contacts
|
4539.35 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Pavlov. Cond. index score
|
1769.33 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Pavlov. Cond. latency score
|
1841.19 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Pavlov. Cond. lever contacts
|
3029.77 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Pavlov. Cond. magazine entry number
|
2162.19 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Pavlov. Cond. intertrial magazine entries
|
524.02 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Pavlov. Cond. lever-magazine prob. diff.
|
1940.27 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Pavlov. Cond. response bias
|
2606.99 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Bone: apparent density
|
22268.03 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Bone surface
|
25125.6 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Bone volume
|
33831.7 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Bone: connectivity density
|
7098.55 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Bone: cortical apparent density
|
45266.32 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Bone: cortical area
|
224.76 |
27 |
2 |
8.3 |
0.66 |
2.94e-12 |
Ablim1
Afap1l2
Cacul1
Casp7
Ces2c
Dclre1a
Dennd10
Fhip2a
Grk5
Habp2
LOC102547573
LOC120100061
LOC120100065
LOC120100068
LOC134485255
LOC134485267
Nanos1
Nhlrc2
Nrap
Plekhs1
Prlhr
Rab11fip2
Retreg2
Sfxn4
Trub1
Vti1a
Zfp933l1
|
|
Bone: cortical porosity
|
1837.29 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Bone: cortical porosity
|
4807.78 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Bone: cortical thickness
|
279.97 |
5 |
1 |
4.2 |
-1 |
2.22e-05 |
Cacul1
LOC120100068
Rab11fip2
Retreg2
Sfxn4
|
|
Bone: cortical thickness
|
12190.6 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Bone: cortical tissue density
|
9699.25 |
4 |
1 |
4.2 |
-0.89 |
4.12e-02 |
Grk5
LOC134485267
Retreg2
Sfxn4
|
|
Bone: elastic displacement
|
29626.25 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Bone: elastic work
|
1494.59 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Bone: endosteal estimation
|
35257.61 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Bone: endosteal perimeter
|
22353.48 |
2 |
1 |
4.2 |
0 |
1.00e+00 |
LOC134485258
Retreg2
|
|
Bone: final force
|
649.79 |
4 |
1 |
4.2 |
0.83 |
1.60e-03 |
Cacul1
Grk5
Retreg2
Sfxn4
|
|
Bone: final moment
|
106.26 |
4 |
1 |
4.2 |
0.93 |
3.48e-05 |
Cacul1
Grk5
Retreg2
Sfxn4
|
|
Bone: marrow area
|
37067.95 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Bone: maximum diameter
|
402.81 |
23 |
1 |
4.2 |
0.73 |
1.12e-11 |
Ablim1
Acsl5
Afap1l2
Cacul1
Casp7
Dclre1a
Fhip2a
Gfra1
Habp2
LOC102547573
LOC102556644
LOC120100061
LOC120100065
LOC134485255
LOC134485258
Nhlrc2
Nrap
Plekhs1
Retreg2
Tcf7l2
Trub1
Vti1a
Zdhhc6
|
|
Bone: maximum force
|
4944.16 |
3 |
1 |
4.2 |
0.83 |
1.94e-02 |
Cacul1
Grk5
Retreg2
|
|
Bone: maximum moment
|
3098.15 |
4 |
1 |
4.2 |
0.84 |
9.42e-03 |
Cacul1
Grk5
Retreg2
Sfxn4
|
|
Bone: minimum diameter
|
17196.64 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Bone: periosteal estimation
|
856.45 |
17 |
1 |
4.2 |
0.63 |
3.96e-06 |
Cacul1
Casp7
Ces2c
Dennd10
Gpam
Grk5
Habp2
LOC120100061
LOC134485255
LOC134485267
Nhlrc2
Prlhr
Rab11fip2
Retreg2
Sfxn4
Tcf7l2
Vti1a
|
|
Bone: periosteal perimeter
|
282.67 |
37 |
2 |
8.3 |
0.58 |
1.55e-15 |
Ablim1
Acsl5
Afap1l2
Atrnl1
Cacul1
Casp7
Ccdc186
Ces2c
Dclre1a
Dennd10
Fhip2a
Gfra1
Gpam
Grk5
Gucy2g
Habp2
LOC102547573
LOC102551125
LOC102556644
LOC120097692
LOC120100061
LOC120100065
LOC134485255
LOC134485257
LOC134485258
LOC134485267
Nhlrc2
Nrap
Plekhs1
Rab11fip2
Retreg2
Sfxn4
Tcf7l2
Trub1
Vti1a
Zdhhc6
Zfp933l1
|
|
Bone: post-yield work
|
50847.58 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Bone: stiffness
|
35564.91 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Bone: tissue strength
|
12264.67 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Bone: trabecular number
|
41138.39 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Bone: trabecular spacing
|
31287.85 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Bone: trabecular thickness
|
2229.65 |
6 |
1 |
4.2 |
0.79 |
4.34e-04 |
Cacul1
Grk5
LOC134485267
Rab11fip2
Retreg2
Sfxn4
|
|
Bone: trabecular tissue density
|
53620.06 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Breakpoint value in progressive ratio session
|
112428.06 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Distance traveled before self-admin
|
134013.73 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Distance traveled after self-admin
|
100820.36 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Delta time in closed arm before/after self-admin
|
1271.86 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Delta time in open arm before/after self-admin
|
54811.83 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Diff in mean of infusions in LGA sessions
|
108490.77 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Extinction: sum of active levers before priming
|
91573.47 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Active lever presses in extinction session 6
|
3117.3 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Delta distance traveled before/after self-admin
|
12434.77 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Delta ambulatory time before/after self-admin
|
96034.01 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Time in closed arm before self-admin
|
88239.28 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Time in closed arm after self-admin
|
19471.75 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Time in open arm before self-admin
|
34587.99 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Time in open arm after self-admin
|
1894.09 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Sum of active levers in priming session hrs 5-6
|
3877.89 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Time to tail flick, vehicle, before self-admin
|
51335.42 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Time to tail flick, vehicle, after self-admin
|
59148.38 |
2 |
1 |
4.2 |
0 |
1.00e+00 |
Gmppa
Retreg2
|
|
Time to tail flick, test, before self-admin
|
32803.11 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Time to tail flick, test, after self-admin
|
5828.87 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Delta time to tail flick, vehicle, before/after SA
|
36533.83 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Delta time to tail flick, test, before/after SA
|
56832.34 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Ambulatory time before self-admin
|
171079.71 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Ambulatory time after self-admin
|
88230.17 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Total heroin consumption
|
961.5 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Area under the delay curve
|
81605.23 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Delay disc. indifference point, 0s delay
|
9477.47 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Delay disc. indifference point, 16s delay
|
74803.61 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Delay disc. indifference point, 2s delay
|
825.84 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Delay disc. indifference point, 24s delay
|
99983.64 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Delay disc. indifference point, 4s delay
|
2525.7 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Delay disc. indifference point, 8s delay
|
101423.99 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Delay discount exponential model param
|
60214.95 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Delay discount hyperbolic model param
|
45715.54 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Fecal boli incidents, locomotor time 1
|
150160.89 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Change in fecal boli incidents, locomotor task
|
23481.3 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Time >=10cm from walls, locomotor time 1
|
49255.61 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Bouts of movement, locomotor time 1
|
41590.88 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Total resting periods, locomotor time 1
|
11802.64 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Rest time, locomotor task time 1
|
75951.88 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Distance moved, locomotor task time 1
|
159283.67 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Vertical activity count, locomotor time 1
|
219061.96 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Time >=10cm from walls, locomotor time 2
|
161986.9 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Bouts of movement, locomotor time 2
|
174298.32 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Total resting periods, locomotor time 2
|
190079.87 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Rest time, locomotor task time 2
|
83778.18 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Distance moved, locomotor task time 2
|
34268.85 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Vertical activity count, locomotor time 2
|
138414.05 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Weight adjusted by age
|
161973.18 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Seeking ratio, delayed vs. immediate footshock
|
49395.56 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Locomotion in novel chamber
|
16301.47 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Locomotion in novel chamber post-restriction
|
154642.04 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Progressive ratio
|
157894.89 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Food seeking constrained by brief footshock
|
57.53 |
13 |
1 |
4.2 |
-0.8 |
7.41e-39 |
Asxl2
Dnmt3a
Drc1
Dtnb
Efr3b
Garem2
Hadha
Hadhb
Kif3c
Otof
Rab10
Retreg2
Yipf4l1
|
|
Seeking ratio, punishment vs. effort
|
118281.2 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Run reversals in cocaine runway, females
|
37174.37 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Run reversals in cocaine runway, males
|
29861.56 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Latency to leave start box in cocaine runway
|
58575.8 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Latency to leave start box in cocaine runway, F
|
28379.24 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Latency to leave start box in cocaine runway, M
|
7140.1 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Co content in liver
|
4721.74 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Cu content in liver
|
162.43 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Fe content in liver
|
5656.2 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
K content in liver
|
6593.53 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Mg content in liver
|
24493.99 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Mn content in liver
|
4374.36 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Na content in liver
|
16641.94 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Rb content in liver
|
428.45 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|
|
Zn content in liver
|
15755.62 |
1 |
1 |
4.2 |
0 |
1.00e+00 |
Retreg2
|