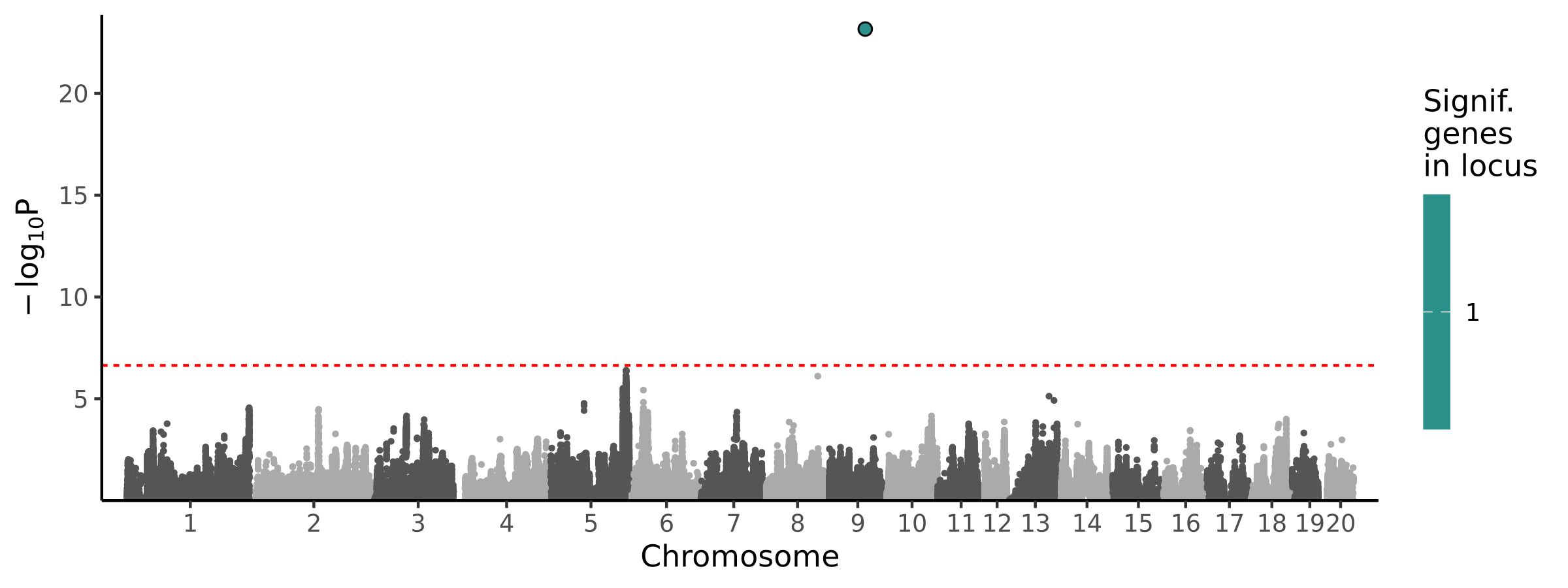
|
BMI with tail
|
355.8 |
4 |
4 |
36.4 |
0.98 |
3.49e-08 |
Cacul1
Grk5
Retreg2
Sfxn4
|
|
BMI without tail
|
11092.69 |
5 |
5 |
45.5 |
-0.99 |
9.07e-10 |
Cacul1
Ccny
Grk5
Retreg2
Sfxn4
|
|
Body weight
|
32.83 |
4 |
3 |
27.3 |
0.01 |
9.82e-01 |
Cacul1
Grk5
Retreg2
Sfxn4
|
|
Epididymis fat weight
|
8369.33 |
5 |
5 |
45.5 |
-0.98 |
3.01e-09 |
Cacul1
Ccny
Grk5
Retreg2
Sfxn4
|
|
Glucose
|
303.08 |
4 |
1 |
9.1 |
1 |
1.45e-08 |
Cacul1
Grk5
Retreg2
Sfxn4
|
|
Left kidney weight
|
10872.68 |
5 |
4 |
36.4 |
0.99 |
8.23e-12 |
Cacul1
Ccny
Grk5
Retreg2
Sfxn4
|
|
Right kidney weight
|
8998.21 |
5 |
5 |
45.5 |
0.99 |
5.80e-12 |
Cacul1
Ccny
Grk5
Retreg2
Sfxn4
|
|
Tail length
|
46066.56 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Length with tail
|
514.51 |
4 |
1 |
9.1 |
-0.98 |
3.75e-08 |
Cacul1
Grk5
Retreg2
Sfxn4
|
|
Length without tail
|
9952.26 |
5 |
2 |
18.2 |
0.99 |
1.82e-11 |
Cacul1
Ccny
Grk5
Retreg2
Sfxn4
|
|
Liver weight, left
|
980.89 |
6 |
5 |
45.5 |
0.99 |
6.82e-14 |
Cacul1
Ccny
Grk5
Retreg2
Sfxn4
Tnfrsf9
|
|
Liver weight, right
|
31.51 |
5 |
1 |
9.1 |
0.89 |
2.17e-04 |
Cacul1
Ccny
Grk5
Retreg2
Sfxn4
|
|
Parametrial fat weight
|
85.3 |
5 |
4 |
36.4 |
-0.65 |
1.58e-02 |
Cacul1
Ccny
Grk5
Retreg2
Sfxn4
|
|
Retroperitoneal fat weight
|
1379.92 |
4 |
4 |
36.4 |
-0.95 |
1.87e-06 |
Cacul1
Grk5
Retreg2
Sfxn4
|
|
Intraocular pressure
|
40071.91 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Extensor digitorum longus weight
|
1562.7 |
3 |
1 |
9.1 |
0 |
1.00e+00 |
Grk5
Retreg2
Sfxn4
|
|
Soleus weight
|
40.92 |
2 |
1 |
9.1 |
0 |
1.00e+00 |
Cacul1
Retreg2
|
|
Tibialis anterior weight
|
4881.94 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Tibia length
|
3259.43 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Number of licking bursts
|
157119.65 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Food consumed during 24 hour testing period
|
53104.21 |
2 |
1 |
9.1 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Times rat made contact with spout
|
305.36 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Mean time between licks in bursts
|
99755.05 |
2 |
1 |
9.1 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Mean num. licks in bursts
|
71302.54 |
2 |
1 |
9.1 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Std. dev. time between licks in bursts
|
94042.02 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Water consumed over 24 hours
|
51711.02 |
2 |
1 |
9.1 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Patch foraging indifference point 0 sec
|
38337.41 |
2 |
1 |
9.1 |
0 |
1.00e+00 |
Cacul1
Retreg2
|
|
Patch foraging indifference point AUC
|
66593.63 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Indifference point function ln k
|
71038.09 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Indifference point function log k
|
71038.09 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Patch foraging total patch changes 0 sec
|
10936.72 |
4 |
1 |
9.1 |
-0.99 |
6.41e-09 |
Cacul1
Grk5
Retreg2
Sfxn4
|
|
Patch foraging total patch changes 12 sec
|
5502.59 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Patch foraging total patch changes 18 sec
|
58887.78 |
2 |
1 |
9.1 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Patch foraging total patch changes 24 sec
|
83555.1 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Patch foraging total patch changes 6 sec
|
16352.42 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Patch foraging time to switch 0 sec
|
156618.53 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Patch foraging water rate 0 sec
|
148807.58 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Patch foraging water rate 12 sec
|
80953.12 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Patch foraging water rate 18 sec
|
51643.12 |
2 |
1 |
9.1 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Patch foraging water rate 24 sec
|
2357.13 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Patch foraging water rate 6 sec
|
2028.63 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Locomotor activity
|
35.04 |
2 |
1 |
9.1 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Locomotor testing distance
|
44778.43 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Locomotor testing rearing
|
152414.81 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Light reinforcement 1
|
77669.42 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Light reinforcement 2
|
54020.7 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Reaction time number correct
|
76747.74 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Reaction time mean minus median
|
102906.71 |
2 |
1 |
9.1 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Reaction time mean minus median AUC
|
208396.98 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Reaction time num false alarms
|
28557.11 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Reaction time num false alarms AUC
|
53265.52 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Reaction time trials correct on left
|
76747.74 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Reaction time trials on left
|
75015.72 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Reaction time mean
|
94380.53 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Reaction time mean AUC
|
56674.13 |
2 |
1 |
9.1 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Median of all reaction times
|
31417.12 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Reaction time omissions
|
34267.91 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Reaction time false alarm rate
|
891.26 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Reaction time premature initiation rate
|
18442.81 |
2 |
1 |
9.1 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Reaction time premature initiations
|
69362.16 |
2 |
1 |
9.1 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Std. dev. reaction times
|
149487.71 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Reaction time trials completed
|
75015.72 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Reaction time trials AUC
|
75869.15 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Social response proportion
|
35302.65 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Social responses
|
2856.43 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Social time
|
44238.34 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Conditioned locomotion
|
3044.63 |
2 |
1 |
9.1 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Cocaine response after cond. corrected
|
32913.12 |
2 |
1 |
9.1 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Cocaine response after cond. not corrected
|
64247.35 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Cocaine response before conditioning
|
1409.02 |
2 |
1 |
9.1 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Saline control response
|
12962.78 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Condit. Reinf. active minus inactive responses
|
53410.48 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Condit. Reinf. active-inactive response ratio
|
91582.24 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Condit. Reinf. active responses
|
9691.24 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Condit. Reinf. inactive responses
|
42106.18 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Condit. Reinf. lever presses
|
15873.43 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Condit. Reinf. lever reinforcers received
|
2000.77 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Pavlov. Cond. lever latency
|
10912.53 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Pavlov. Cond. magazine entry latency
|
24714.14 |
2 |
1 |
9.1 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Pavlov. Cond. change in total contacts
|
114781.94 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Pavlov. Cond. index score
|
23998.4 |
2 |
1 |
9.1 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Pavlov. Cond. latency score
|
29242.48 |
2 |
1 |
9.1 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Pavlov. Cond. lever contacts
|
22069.78 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Pavlov. Cond. magazine entry number
|
16773.11 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Pavlov. Cond. intertrial magazine entries
|
104933.78 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Pavlov. Cond. lever-magazine prob. diff.
|
44871.67 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Pavlov. Cond. response bias
|
5169.79 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Conditioned reinforcement - actives
|
95753.16 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Conditioned reinforcement - inactives
|
77525.72 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Conditioned locomotion
|
20734.34 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Intermittent access intake day 1-15 change
|
105306.64 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Intermittent access intake escalation
|
209543.53 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Intermittent access intake escalation 2
|
54050.4 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Intermitt. access day 1 inactive lever presses
|
118272.07 |
2 |
1 |
9.1 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Intermitt. access day 15 inactive lever presses
|
208241.66 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Intermitt. access escalation Index
|
92502.05 |
2 |
1 |
9.1 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Intermittent access day 1 total infusions
|
77765.06 |
2 |
1 |
9.1 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Intermittent access day 15 total infusions
|
65747.49 |
2 |
1 |
9.1 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Intermittent access terminal intake (last 3 days)
|
83205.14 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Intermittent access total infusions
|
345.81 |
2 |
1 |
9.1 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Intermittent access day 1 locomotion
|
52247.41 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Intermittent access day 15 total locomotion
|
88947.95 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Intermittent access total locomotion
|
711.59 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Intermittent access standard deviation
|
100979.14 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Cocaine induced anxiety
|
80149.68 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Post-drug Anxiety
|
20460.83 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Baseline Anxiety
|
51769.43 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Lifetime Intake
|
3437.35 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Incentive Sensitization - Responses
|
81797.54 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Incentive Sensitization - Breakpoint
|
71404.4 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Progressive ratio test 1 active lever presses
|
49966.26 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Progressive ratio test 1 breakpoint
|
48151.76 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Progressive ratio test 1 inactive lever presses
|
75556.29 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Progressive ratio test 2 active lever presses
|
28633.56 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Progressive ratio test 2 breakpoint
|
17520.06 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Progressive ratio test 2 inactive lever presses
|
82060.82 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Total sessions with >9 infusions
|
4022.32 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Short access day 1 total inactive lever presses
|
57203.57 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Short access day 10 total inactive lever presses
|
108624.06 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Short access day 1 total infusions
|
55866.13 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Short access day 10 total infusions
|
38150.26 |
2 |
1 |
9.1 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Short access total infusions
|
17026.24 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Short access day 1 locomotion
|
42054.78 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Short access day 10 total locomotion
|
90804.25 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Short access total locomotion
|
68266.59 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Compulsive drug intake
|
32724.39 |
2 |
1 |
9.1 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
One hour access (0.3 mA shock)
|
4858.65 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
One hour access (shock baseline)
|
70135.77 |
2 |
1 |
9.1 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Number of responses in last shaping day
|
25706.97 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Context. condit. distance diff. score
|
970.59 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Locomotion velocity, session 1
|
14615.25 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Locomotion distance, session 1
|
2213.01 |
2 |
1 |
9.1 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Locomotion velocity, session 2
|
9358 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Locomotion distance, session 2
|
1206.44 |
2 |
1 |
9.1 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Locomotion velocity, session 3
|
2332.53 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Locomotion distance, session 3
|
16615.78 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Stereotopy head waving bouts, day 3
|
5866.4 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Stereotopy head waving duration, day 3
|
12131.92 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Locomotion velocity, session 7
|
1310.17 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Locomotion distance, session 7
|
3744.54 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Stereotopy head waving bouts, day 7
|
10466.92 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Stereotopy head waving duration, day 7
|
8847.36 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Locomotion velocity, session 8
|
1892.98 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Locomotion distance, session 8
|
229.73 |
2 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
Tnfrsf9
|
|
Degree of sensitization distance
|
15309.16 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Degree of sensitization stereotypy
|
8496.38 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Condit. Reinf. active minus inactive responses
|
2066.34 |
2 |
1 |
9.1 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Condit. Reinf. active-inactive response ratio
|
1097.66 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Condit. Reinf. active responses
|
5839.15 |
2 |
1 |
9.1 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Condit. Reinf. inactive responses
|
2430.04 |
6 |
1 |
9.1 |
-0.99 |
3.49e-06 |
Ctnnbip1
LOC134487042
Nmnat1
Rbp7
Retreg2
Tnfrsf9
|
|
Incentive salience index mean
|
7397.37 |
2 |
1 |
9.1 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Condit. Reinf. lever presses
|
21433.81 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Time in familiar zone, hab. session 1
|
41873.03 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Time in novel zone, hab. session 1
|
35522.88 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Total zone transitions, hab. session 1
|
1818.53 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Total locomotion distance, hab. session 1
|
849.35 |
2 |
1 |
9.1 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Locomotion velocity, hab. session 1
|
571.33 |
2 |
1 |
9.1 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Time in familiar zone, hab. session 2
|
23379.25 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Time in novel zone, hab. session 2
|
24924.95 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Total zone transitions, hab. session 2
|
280.53 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Total locomotion distance, hab. session 2
|
96.8 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Locomotion velocity, hab. session 2
|
77.91 |
2 |
1 |
9.1 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Novel to familiar place preference ratio
|
27759.88 |
2 |
1 |
9.1 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Novelty place preference
|
54383.96 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Time in novel zone, NPP test
|
40844.77 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Total zone transitions, NPP test
|
8230.03 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Total locomotion distance, NPP test
|
3566.83 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Locomotion velocity, NPP test
|
2031.76 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Pavlov. Cond. lever latency
|
2195.61 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Pavlov. Cond. magazine entry latency
|
8.04 |
1 |
0 |
0 |
0 |
1.00e+00 |
Ccny
|
|
Pavlov. Cond. change in total contacts
|
4539.35 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Pavlov. Cond. index score
|
1769.33 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Pavlov. Cond. latency score
|
1841.19 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Pavlov. Cond. lever contacts
|
3029.77 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Pavlov. Cond. magazine entry number
|
1086.53 |
2 |
1 |
9.1 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Pavlov. Cond. intertrial magazine entries
|
524.02 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Pavlov. Cond. lever-magazine prob. diff.
|
1940.27 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Pavlov. Cond. response bias
|
2606.99 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Bone: apparent density
|
4458.81 |
2 |
1 |
9.1 |
-0.99 |
8.24e-04 |
Cacul1
Retreg2
|
|
Bone surface
|
6285.49 |
2 |
1 |
9.1 |
-0.99 |
5.08e-03 |
Cacul1
Retreg2
|
|
Bone volume
|
3389.57 |
4 |
1 |
9.1 |
-0.99 |
1.54e-07 |
Cacul1
Grk5
Retreg2
Sfxn4
|
|
Bone: connectivity density
|
7098.55 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Bone: cortical apparent density
|
3780.1 |
4 |
1 |
9.1 |
0.98 |
7.56e-09 |
Cacul1
Grk5
Retreg2
Sfxn4
|
|
Bone: cortical area
|
1591.32 |
4 |
2 |
18.2 |
-0.97 |
8.36e-08 |
Cacul1
Grk5
Retreg2
Sfxn4
|
|
Bone: cortical porosity
|
1837.29 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Bone: cortical porosity
|
4807.78 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Bone: cortical thickness
|
143.77 |
4 |
1 |
9.1 |
0.99 |
1.63e-10 |
Cacul1
Grk5
Retreg2
Sfxn4
|
|
Bone: cortical thickness
|
12190.6 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Bone: cortical tissue density
|
4046.36 |
4 |
1 |
9.1 |
0.98 |
7.68e-09 |
Cacul1
Grk5
Retreg2
Sfxn4
|
|
Bone: elastic displacement
|
29626.25 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Bone: elastic work
|
1494.59 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Bone: endosteal estimation
|
35257.61 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Bone: endosteal perimeter
|
44697.39 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Bone: final force
|
595.9 |
4 |
1 |
9.1 |
-0.97 |
3.82e-07 |
Cacul1
Grk5
Retreg2
Sfxn4
|
|
Bone: final moment
|
97.82 |
4 |
1 |
9.1 |
-0.89 |
1.21e-04 |
Cacul1
Grk5
Retreg2
Sfxn4
|
|
Bone: marrow area
|
37067.95 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Bone: maximum diameter
|
2040.38 |
4 |
1 |
9.1 |
-0.98 |
1.51e-08 |
Cacul1
Grk5
Retreg2
Sfxn4
|
|
Bone: maximum force
|
3148.85 |
4 |
1 |
9.1 |
-0.98 |
3.90e-08 |
Cacul1
Grk5
Retreg2
Sfxn4
|
|
Bone: maximum moment
|
2067.73 |
4 |
1 |
9.1 |
-0.98 |
1.92e-08 |
Cacul1
Grk5
Retreg2
Sfxn4
|
|
Bone: minimum diameter
|
1440.3 |
4 |
1 |
9.1 |
-0.98 |
2.95e-08 |
Cacul1
Grk5
Retreg2
Sfxn4
|
|
Bone: periosteal estimation
|
3191.26 |
4 |
1 |
9.1 |
-0.98 |
1.10e-08 |
Cacul1
Grk5
Retreg2
Sfxn4
|
|
Bone: periosteal perimeter
|
3608.45 |
4 |
1 |
9.1 |
-0.98 |
1.09e-08 |
Cacul1
Grk5
Retreg2
Sfxn4
|
|
Bone: post-yield work
|
50847.58 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Bone: stiffness
|
35564.91 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Bone: tissue strength
|
12264.67 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Bone: trabecular number
|
41138.39 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Bone: trabecular spacing
|
31287.85 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Bone: trabecular thickness
|
2783.53 |
4 |
1 |
9.1 |
-0.98 |
2.09e-08 |
Cacul1
Grk5
Retreg2
Sfxn4
|
|
Bone: trabecular tissue density
|
26819.29 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Breakpoint value in progressive ratio session
|
56221.29 |
2 |
1 |
9.1 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Distance traveled before self-admin
|
67038.76 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Distance traveled after self-admin
|
50469.31 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Delta time in closed arm before/after self-admin
|
1030.46 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Delta time in open arm before/after self-admin
|
27441.91 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Diff in mean of infusions in LGA sessions
|
108490.77 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Extinction: sum of active levers before priming
|
45875 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Active lever presses in extinction session 6
|
1673.08 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Delta distance traveled before/after self-admin
|
6292.84 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Delta ambulatory time before/after self-admin
|
48052.09 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Time in closed arm before self-admin
|
44143.36 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Time in closed arm after self-admin
|
9901.19 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Time in open arm before self-admin
|
17365.6 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Time in open arm after self-admin
|
981.98 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Sum of active levers in priming session hrs 5-6
|
3877.89 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Time to tail flick, vehicle, before self-admin
|
25931.02 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Time to tail flick, vehicle, after self-admin
|
59159.07 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Time to tail flick, test, before self-admin
|
16410.58 |
2 |
1 |
9.1 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Time to tail flick, test, after self-admin
|
2994.46 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Delta time to tail flick, vehicle, before/after SA
|
18449.53 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Delta time to tail flick, test, before/after SA
|
28423.85 |
2 |
1 |
9.1 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Ambulatory time before self-admin
|
85572.63 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Ambulatory time after self-admin
|
44153.3 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Total heroin consumption
|
547.18 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Area under the delay curve
|
40810.48 |
2 |
1 |
9.1 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Delay disc. indifference point, 0s delay
|
9477.47 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Delay disc. indifference point, 16s delay
|
37437.33 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Delay disc. indifference point, 2s delay
|
825.84 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Delay disc. indifference point, 24s delay
|
50025 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Delay disc. indifference point, 4s delay
|
1298.29 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Delay disc. indifference point, 8s delay
|
101423.99 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Delay discount exponential model param
|
60214.95 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Delay discount hyperbolic model param
|
45715.54 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Fecal boli incidents, locomotor time 1
|
50057.92 |
3 |
1 |
9.1 |
0 |
1.00e+00 |
Ccny
Retreg2
Tnfrsf9
|
|
Change in fecal boli incidents, locomotor task
|
11821.21 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Time >=10cm from walls, locomotor time 1
|
24655.43 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Bouts of movement, locomotor time 1
|
20842.46 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Total resting periods, locomotor time 1
|
5936.78 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Rest time, locomotor task time 1
|
37983.55 |
2 |
1 |
9.1 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Distance moved, locomotor task time 1
|
79651.82 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Vertical activity count, locomotor time 1
|
219061.96 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Time >=10cm from walls, locomotor time 2
|
80995.71 |
2 |
1 |
9.1 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Bouts of movement, locomotor time 2
|
87164.71 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Total resting periods, locomotor time 2
|
95051.56 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Rest time, locomotor task time 2
|
41900.71 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Distance moved, locomotor task time 2
|
17217.33 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Vertical activity count, locomotor time 2
|
69238.05 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Weight adjusted by age
|
32412.75 |
5 |
2 |
18.2 |
-0.99 |
5.02e-04 |
Cacul1
Ccny
Grk5
Retreg2
Sfxn4
|
|
Seeking ratio, delayed vs. immediate footshock
|
8241.51 |
4 |
2 |
18.2 |
-1 |
2.28e-06 |
Ccny
Grk5
Retreg2
Sfxn4
|
|
Locomotion in novel chamber
|
8161.06 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Locomotion in novel chamber post-restriction
|
77324.49 |
2 |
1 |
9.1 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Progressive ratio
|
78975.01 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Food seeking constrained by brief footshock
|
1230.38 |
4 |
1 |
9.1 |
0.99 |
1.58e-04 |
Cacul1
Grk5
Retreg2
Sfxn4
|
|
Seeking ratio, punishment vs. effort
|
59148.59 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Run reversals in cocaine runway, females
|
18598.36 |
2 |
1 |
9.1 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Run reversals in cocaine runway, males
|
14942.17 |
2 |
1 |
9.1 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Latency to leave start box in cocaine runway
|
29317.55 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Latency to leave start box in cocaine runway, F
|
28379.24 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Latency to leave start box in cocaine runway, M
|
3675.36 |
2 |
2 |
18.2 |
0 |
1.00e+00 |
Ccny
Retreg2
|
|
Co content in liver
|
4721.74 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Cu content in liver
|
162.43 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Fe content in liver
|
5656.2 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
K content in liver
|
6593.53 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Mg content in liver
|
24493.99 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Mn content in liver
|
4374.36 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Na content in liver
|
16641.94 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Rb content in liver
|
428.45 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|
|
Zn content in liver
|
15755.62 |
1 |
1 |
9.1 |
0 |
1.00e+00 |
Retreg2
|