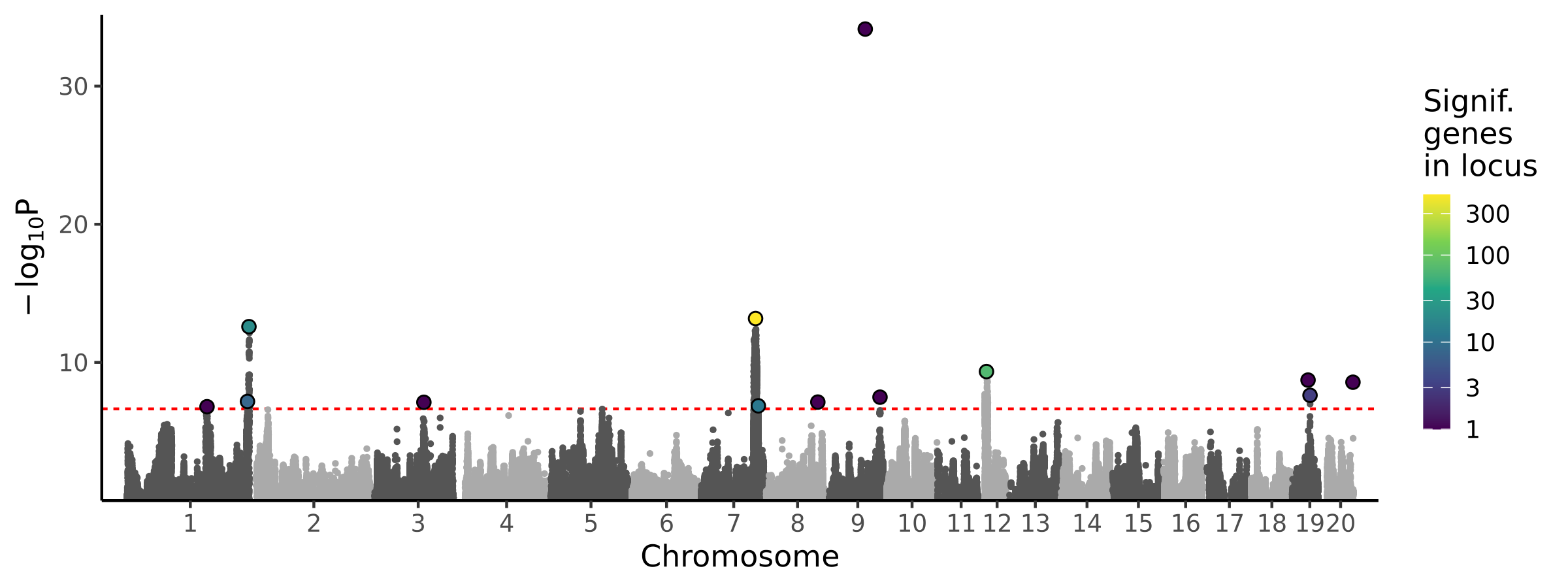
|
BMI with tail
|
24.77 |
28 |
18 |
58.1 |
0.96 |
9.15e-44 |
Cacul1
Casp7
Ces2c
Dennd10
Emx2os
Fam204a
Grk5
Klhdc8b
Lmf2
LOC102549998
LOC102550729
LOC103694460
LOC120100064
LOC120100068
Mapk11
Nanos1
Plekha7
Plxnb2
Ppp6r2
Prdx3
Prlhr
Rab11fip2
Selenoo
Sfxn4
Tafa5
Tymp
Zfand2b
Zfp950
|
|
BMI without tail
|
20.81 |
16 |
15 |
48.4 |
0.88 |
9.23e-20 |
Cacul1
Ces2c
Dennd10
Emx2os
Fam204a
Grk5
Klhdc8b
LOC102550729
Nanos1
Plekha7
Prdx3
Prlhr
Rab11fip2
Sfxn4
Zfand2b
Zfp950
|
|
Body weight
|
19.53 |
50 |
43 |
138.7 |
0.98 |
5.35e-131 |
Ablim1
Afap1l2
Atrnl1
Brca2
Cacul1
Casp7
Cers4
Ces2c
Dennd10
Elavl1
Emx2os
Fam204a
Fry
Grk5
Hmgb1
Hsph1
Insr
Katnal1
Klhdc8b
LOC102548389
LOC102549089
LOC102549494
LOC102550729
LOC102553253
LOC103694460
LOC108352411
LOC120095830
LOC120095871
LOC120100064
LOC120100068
Mcoln1
Medag
N4bp2l1
N4bp2l2
Nanos1
Pet100
Plekha7
Prdx3
Prlhr
Rab11fip2
Rxfp2
Sfxn4
Slc7a1
Snapc2
Stard13
Stxbp2
Tafa5
Trub1
Xab2
Zfp950
|
|
Epididymis fat weight
|
32.47 |
21 |
15 |
48.4 |
0.87 |
1.05e-21 |
Ablim1
Afap1l2
Atrnl1
Cacul1
Casp7
Ces2c
Dennd10
Emx2os
Fam204a
Grk5
LOC102550729
LOC120100064
LOC120100068
Nanos1
Prdx3
Prlhr
Rab11fip2
Sfxn4
Trub1
Zfand2b
Zfp950
|
|
Heart weight
|
22.96 |
5 |
1 |
3.2 |
0.99 |
5.14e-05 |
Cacul1
Emx2os
Klhdc8b
Sfxn4
Zfand2b
|
|
Left kidney weight
|
13.88 |
155 |
136 |
438.7 |
1 |
0.00e+00 |
A4galt
Ankrd54
Arfgap3
Arhgap8
Atrnl1
Atxn10
Baiap2l2
Bik
Brca2
Cacna1i
Cacul1
Casp7
Cbx7
Cby1
Ccdc134
Cdpf1
Celsr1
Cenpm
Cerk
Cers4
Ces2c
Csdc2
Cyb5r3
Cyp2d1
Cyp2d3
Cyp2d4
Cyp2d5
Dennd10
Desi1
Efcab6
Elavl1
Emx2os
Fam118a
Fam204a
Fbln1
Fry
Gramd4
Grap2
Grk5
Gtse1
Hmgb1
Hsph1
Insr
Ist1
Itpka
Josd1
Katnal1
Klhdc8b
L3mbtl2
LOC100362109
LOC102547313
LOC102548389
LOC102549089
LOC102549144
LOC102549494
LOC102549885
LOC102550729
LOC102553253
LOC102554082
LOC103692950
LOC103694460
LOC108351520
LOC108352411
LOC120093690
LOC120093696
LOC120093702
LOC120093706
LOC120095830
LOC120095871
LOC120100064
LOC120100068
Mcat
Mchr1
Mcoln1
Medag
Mei1
Mgat3
Mief1
Mpped1
Mrtfa
N4bp2l1
N4bp2l2
Nanos1
Ndufa6
Ndufb11b
Nfam1
Nptxr
Nup50
Pacsin2
Parvb
Parvg
Pet100
Pheta2
Phf21b
Phf5a
Pick1
Plekha7
Pmfbp1
Pmm1
Pnpla3
Poldip3
Polr3h
Ppara
Prdx3
Prlhr
Prr5
Rab11fip2
Rbx1
RGD1304694
RGD1359634
Rrp7a
Rxfp2
Samm50
Scube1
Septin3
Serhl2
Sfxn4
Shisa8
Shisal1
Slc16a8
Slc25a17
Smdt1
Smim45
Snapc2
Snu13
Srebf2
St13
Stard13
Stxbp2
Sult4a1
Sun2
Syngr1
Tab1
Tafa5
Tbc1d22a
Tcf20
Tef
Tmem184b
Tnrc6b
Tob2
Trmu
Tspo
Ttc38
Ttll1
Ttll12
Ube2s-ps3
Wbp2nl
Wnt7b
Xab2
Xpnpep3
Xrcc6
Zc3h7b
Zdhhc1
Zfand2b
Zfp950
|
|
Tail length
|
10.99 |
18 |
15 |
48.4 |
0.77 |
7.85e-15 |
Brca2
Cers4
Elavl1
Insr
Katnal1
Klhdc8b
LOC102548389
LOC102549089
LOC120095871
Mcoln1
N4bp2l1
N4bp2l2
Pet100
Snapc2
Stard13
Stxbp2
Xab2
Zfand2b
|
|
Length with tail
|
16.43 |
30 |
25 |
80.6 |
0.94 |
1.96e-50 |
Brca2
Cacul1
Cers4
Elavl1
Fry
Hmgb1
Hsph1
Insr
Katnal1
Klhdc8b
LOC102548389
LOC102549089
LOC102549494
LOC102553253
LOC108352411
LOC120095830
LOC120095871
Mcoln1
Medag
N4bp2l1
N4bp2l2
Pet100
Rxfp2
Sfxn4
Slc7a1
Snapc2
Stard13
Stxbp2
Xab2
Zfand2b
|
|
Length without tail
|
14.33 |
30 |
20 |
64.5 |
0.99 |
1.56e-90 |
Brca2
Cacul1
Cers4
Elavl1
Emx2os
Fry
Grk5
Hmgb1
Hsph1
Insr
Katnal1
LOC102548389
LOC102549089
LOC102549494
LOC102553253
LOC108352411
LOC120095830
LOC120095871
Mcoln1
Medag
N4bp2l1
N4bp2l2
Pet100
Prlhr
Rxfp2
Snapc2
Stard13
Stxbp2
Xab2
Zfand2b
|
|
Liver weight, left
|
11.62 |
4 |
0 |
0 |
0.61 |
3.95e-01 |
Cacul1
Grk5
Klhdc8b
Prlhr
|
|
Liver weight, right
|
15.12 |
13 |
7 |
22.6 |
0.82 |
1.99e-06 |
Cacul1
Dennd10
Fam204a
Grk5
LOC102549494
LOC103694460
LOC120095830
Prlhr
Rab11fip2
Rxfp2
Sfxn4
Zfand2b
Zfp950
|
|
Parametrial fat weight
|
23.42 |
16 |
13 |
41.9 |
0.97 |
1.86e-36 |
Cacul1
Ces2c
Dennd10
Emx2os
Fam204a
Grk5
LOC102550729
Nanos1
Plekha7
Prdx3
Prlhr
Rab11fip2
Sfxn4
Zdhhc1
Zfand2b
Zfp950
|
|
Retroperitoneal fat weight
|
22.61 |
48 |
24 |
77.4 |
0.88 |
4.85e-43 |
Ablim1
Afap1l2
Atrnl1
Cacna1i
Cacul1
Casp7
Cbx7
Cby1
Ces2c
Csdc2
Dennd10
Desi1
Emx2os
Fam204a
Grap2
Grk5
Josd1
Klhdc8b
LOC102549144
LOC102550729
LOC103692950
LOC103694460
LOC120093690
LOC120093696
LOC120100064
LOC120100068
Mei1
Mgat3
Nanos1
Nptxr
Pmm1
Polr3h
Prdx3
Prlhr
Rab11fip2
Rbx1
Sfxn4
Slc25a17
Snu13
Sun2
Syngr1
Tab1
Tnrc6b
Trub1
Xpnpep3
Zc3h7b
Zfand2b
Zfp950
|
|
Intraocular pressure
|
38.04 |
2 |
2 |
6.5 |
0 |
1.00e+00 |
LOC103694460
Zfand2b
|
|
Extensor digitorum longus weight
|
6.11 |
19 |
5 |
16.1 |
1 |
2.64e-77 |
Brca2
Cers4
Elavl1
Fry
Insr
LOC102548389
LOC102549089
LOC102549494
LOC103694460
LOC120095871
Mcoln1
N4bp2l1
N4bp2l2
Pet100
Rxfp2
Snapc2
Stard13
Stxbp2
Xab2
|
|
Soleus weight
|
6.36 |
9 |
0 |
0 |
1 |
1.87e-11 |
Ablim1
Afap1l2
Atrnl1
Emx2os
Grk5
Klhdc8b
LOC120100064
LOC120100068
Trub1
|
|
Tibialis anterior weight
|
8.03 |
19 |
14 |
45.2 |
1 |
1.57e-77 |
Brca2
Cers4
Elavl1
Fry
Insr
LOC102548389
LOC102549089
LOC102549494
LOC103694460
LOC120095871
Mcoln1
N4bp2l1
N4bp2l2
Pet100
Rxfp2
Snapc2
Stard13
Stxbp2
Xab2
|
|
Tibia length
|
6.87 |
13 |
1 |
3.2 |
1 |
3.15e-37 |
Brca2
Elavl1
Insr
Klhdc8b
LOC102549494
Mcoln1
N4bp2l1
N4bp2l2
Pet100
Rxfp2
Snapc2
Stard13
Stxbp2
|
|
Number of licking bursts
|
28.51 |
2 |
2 |
6.5 |
0 |
1.00e+00 |
Klhdc8b
Zfand2b
|
|
Food consumed during 24 hour testing period
|
15.78 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zfand2b
|
|
Mean num. licks in bursts
|
18.13 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zfand2b
|
|
Water consumed over 24 hours
|
34.33 |
1 |
1 |
3.2 |
0 |
1.00e+00 |
Zfand2b
|
|
Patch foraging indifference point 0 sec
|
12.38 |
2 |
0 |
0 |
0 |
1.00e+00 |
LOC103694460
Zfand2b
|
|
Patch foraging indifference point AUC
|
24.2 |
1 |
1 |
3.2 |
0 |
1.00e+00 |
Zfand2b
|
|
Indifference point function ln k
|
45.4 |
2 |
2 |
6.5 |
0 |
1.00e+00 |
Klhdc8b
Zfand2b
|
|
Indifference point function log k
|
45.4 |
2 |
2 |
6.5 |
0 |
1.00e+00 |
Klhdc8b
Zfand2b
|
|
Patch foraging total patch changes 0 sec
|
18.06 |
2 |
1 |
3.2 |
0 |
1.00e+00 |
LOC103694460
Zfand2b
|
|
Patch foraging total patch changes 18 sec
|
48.28 |
1 |
1 |
3.2 |
0 |
1.00e+00 |
Zfand2b
|
|
Patch foraging total patch changes 24 sec
|
77.61 |
1 |
1 |
3.2 |
0 |
1.00e+00 |
Zfand2b
|
|
Patch foraging total patch changes 6 sec
|
24.67 |
1 |
1 |
3.2 |
0 |
1.00e+00 |
Zfand2b
|
|
Patch foraging time to switch 0 sec
|
33.47 |
1 |
1 |
3.2 |
0 |
1.00e+00 |
Zfand2b
|
|
Patch foraging water rate 0 sec
|
39.47 |
1 |
1 |
3.2 |
0 |
1.00e+00 |
Zfand2b
|
|
Patch foraging water rate 12 sec
|
69.49 |
1 |
1 |
3.2 |
0 |
1.00e+00 |
Zfand2b
|
|
Patch foraging water rate 24 sec
|
12.61 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zfand2b
|
|
Patch foraging water rate 6 sec
|
35.43 |
1 |
1 |
3.2 |
0 |
1.00e+00 |
Zfand2b
|
|
Locomotor activity
|
35.08 |
2 |
1 |
3.2 |
0 |
1.00e+00 |
Klhdc8b
Zfand2b
|
|
Locomotor testing distance
|
55.86 |
2 |
2 |
6.5 |
0 |
1.00e+00 |
Klhdc8b
Zfand2b
|
|
Locomotor testing rearing
|
99.51 |
1 |
1 |
3.2 |
0 |
1.00e+00 |
Zfand2b
|
|
Light reinforcement 1
|
14.51 |
2 |
1 |
3.2 |
0 |
1.00e+00 |
Zdhhc1
Zfand2b
|
|
Reaction time number correct
|
66.72 |
2 |
2 |
6.5 |
0 |
1.00e+00 |
Klhdc8b
Zfand2b
|
|
Reaction time mean minus median
|
179.03 |
1 |
1 |
3.2 |
0 |
1.00e+00 |
Zfand2b
|
|
Reaction time mean minus median AUC
|
232.47 |
1 |
1 |
3.2 |
0 |
1.00e+00 |
Zfand2b
|
|
Reaction time num false alarms AUC
|
27.43 |
1 |
1 |
3.2 |
0 |
1.00e+00 |
Zfand2b
|
|
Reaction time trials correct on left
|
66.72 |
2 |
2 |
6.5 |
0 |
1.00e+00 |
Klhdc8b
Zfand2b
|
|
Reaction time trials on left
|
63.1 |
2 |
2 |
6.5 |
0 |
1.00e+00 |
Klhdc8b
Zfand2b
|
|
Reaction time mean
|
30.14 |
2 |
2 |
6.5 |
0 |
1.00e+00 |
Zdhhc1
Zfand2b
|
|
Reaction time mean AUC
|
26.7 |
2 |
1 |
3.2 |
0 |
1.00e+00 |
Zdhhc1
Zfand2b
|
|
Median of all reaction times
|
22.05 |
2 |
1 |
3.2 |
0 |
1.00e+00 |
Zdhhc1
Zfand2b
|
|
Reaction time omissions
|
35.15 |
2 |
2 |
6.5 |
0 |
1.00e+00 |
Klhdc8b
Zfand2b
|
|
Reaction time false alarm rate
|
20.24 |
1 |
1 |
3.2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Reaction time premature initiations
|
41.6 |
1 |
1 |
3.2 |
0 |
1.00e+00 |
Zfand2b
|
|
Std. dev. reaction times
|
72.92 |
1 |
1 |
3.2 |
0 |
1.00e+00 |
Zfand2b
|
|
Reaction time trials completed
|
63.1 |
2 |
2 |
6.5 |
0 |
1.00e+00 |
Klhdc8b
Zfand2b
|
|
Reaction time trials AUC
|
64.7 |
2 |
2 |
6.5 |
0 |
1.00e+00 |
Klhdc8b
Zfand2b
|
|
Conditioned locomotion
|
28.4 |
1 |
1 |
3.2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Cocaine response after cond. not corrected
|
16.6 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Cocaine response before conditioning
|
38.66 |
1 |
1 |
3.2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Pavlov. Cond. index score
|
15.43 |
1 |
0 |
0 |
0 |
1.00e+00 |
LOC103694460
|
|
Pavlov. Cond. magazine entry number
|
25.13 |
2 |
1 |
3.2 |
0 |
1.00e+00 |
Klhdc8b
LOC103694460
|
|
Pavlov. Cond. response bias
|
13.13 |
1 |
0 |
0 |
0 |
1.00e+00 |
LOC103694460
|
|
Conditioned reinforcement - actives
|
152.54 |
1 |
1 |
3.2 |
0 |
1.00e+00 |
Zfand2b
|
|
Conditioned reinforcement - inactives
|
76.2 |
2 |
1 |
3.2 |
0 |
1.00e+00 |
Klhdc8b
Zfand2b
|
|
Intermittent access intake day 1-15 change
|
156.77 |
1 |
1 |
3.2 |
0 |
1.00e+00 |
Zfand2b
|
|
Intermittent access intake escalation
|
117.41 |
1 |
1 |
3.2 |
0 |
1.00e+00 |
Zfand2b
|
|
Intermitt. access day 1 inactive lever presses
|
169.63 |
1 |
1 |
3.2 |
0 |
1.00e+00 |
Zfand2b
|
|
Intermitt. access day 15 inactive lever presses
|
162.52 |
2 |
2 |
6.5 |
0 |
1.00e+00 |
Klhdc8b
Zfand2b
|
|
Intermittent access day 1 total infusions
|
71.42 |
1 |
1 |
3.2 |
0 |
1.00e+00 |
Zfand2b
|
|
Intermittent access day 15 total infusions
|
35.39 |
1 |
1 |
3.2 |
0 |
1.00e+00 |
Zfand2b
|
|
Intermittent access terminal intake (last 3 days)
|
24.92 |
2 |
1 |
3.2 |
0 |
1.00e+00 |
Klhdc8b
Zfand2b
|
|
Intermittent access total infusions
|
15.45 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Intermittent access day 1 locomotion
|
26.47 |
1 |
1 |
3.2 |
0 |
1.00e+00 |
Zfand2b
|
|
Intermittent access day 15 total locomotion
|
93.08 |
1 |
1 |
3.2 |
0 |
1.00e+00 |
Zfand2b
|
|
Intermittent access standard deviation
|
229.66 |
2 |
1 |
3.2 |
0 |
1.00e+00 |
Klhdc8b
Zfand2b
|
|
Cocaine induced anxiety
|
125.35 |
1 |
1 |
3.2 |
0 |
1.00e+00 |
Zfand2b
|
|
Baseline Anxiety
|
393.92 |
1 |
1 |
3.2 |
0 |
1.00e+00 |
Zfand2b
|
|
Incentive Sensitization - Responses
|
169.87 |
2 |
2 |
6.5 |
0 |
1.00e+00 |
Zdhhc1
Zfand2b
|
|
Incentive Sensitization - Breakpoint
|
165.51 |
2 |
2 |
6.5 |
0 |
1.00e+00 |
Zdhhc1
Zfand2b
|
|
Progressive ratio test 1 active lever presses
|
26.41 |
1 |
1 |
3.2 |
0 |
1.00e+00 |
Zfand2b
|
|
Progressive ratio test 1 breakpoint
|
19.19 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zfand2b
|
|
Progressive ratio test 1 inactive lever presses
|
16.55 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Progressive ratio test 2 active lever presses
|
67.87 |
2 |
2 |
6.5 |
0 |
1.00e+00 |
Zdhhc1
Zfand2b
|
|
Progressive ratio test 2 breakpoint
|
63.74 |
2 |
1 |
3.2 |
0 |
1.00e+00 |
Zdhhc1
Zfand2b
|
|
Progressive ratio test 2 inactive lever presses
|
63.47 |
1 |
1 |
3.2 |
0 |
1.00e+00 |
Zfand2b
|
|
Total sessions with >9 infusions
|
32.12 |
2 |
1 |
3.2 |
0 |
1.00e+00 |
Klhdc8b
Zdhhc1
|
|
Short access day 1 total inactive lever presses
|
21.93 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zfand2b
|
|
Short access day 10 total inactive lever presses
|
72.35 |
2 |
2 |
6.5 |
0 |
1.00e+00 |
Klhdc8b
Zfand2b
|
|
Short access day 1 total infusions
|
19.14 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zdhhc1
|
|
Short access day 1 locomotion
|
129.19 |
1 |
1 |
3.2 |
0 |
1.00e+00 |
Zfand2b
|
|
Short access day 10 total locomotion
|
432.47 |
1 |
1 |
3.2 |
0 |
1.00e+00 |
Zfand2b
|
|
Short access total locomotion
|
209.96 |
1 |
1 |
3.2 |
0 |
1.00e+00 |
Zfand2b
|
|
Compulsive drug intake
|
68.39 |
1 |
1 |
3.2 |
0 |
1.00e+00 |
Zfand2b
|
|
One hour access (0.3 mA shock)
|
22.58 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zfand2b
|
|
One hour access (shock baseline)
|
14.65 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zdhhc1
|
|
Number of responses in last shaping day
|
67.75 |
1 |
1 |
3.2 |
0 |
1.00e+00 |
Zfand2b
|
|
Locomotion distance, session 1
|
23.98 |
1 |
1 |
3.2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Locomotion distance, session 2
|
33.15 |
1 |
1 |
3.2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Locomotion velocity, session 3
|
17.14 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Locomotion distance, session 8
|
52.78 |
1 |
1 |
3.2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Degree of sensitization distance
|
24.75 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Condit. Reinf. active minus inactive responses
|
35.59 |
1 |
1 |
3.2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Condit. Reinf. active-inactive response ratio
|
25.6 |
1 |
1 |
3.2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Condit. Reinf. active responses
|
18.23 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Incentive salience index mean
|
26.23 |
1 |
1 |
3.2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Time in familiar zone, hab. session 1
|
63.49 |
1 |
1 |
3.2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Total zone transitions, hab. session 1
|
37.38 |
1 |
1 |
3.2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Time in familiar zone, hab. session 2
|
27.1 |
1 |
1 |
3.2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Total zone transitions, hab. session 2
|
69.58 |
1 |
1 |
3.2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Total locomotion distance, hab. session 2
|
19.2 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Locomotion velocity, hab. session 2
|
21.15 |
1 |
1 |
3.2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Novel to familiar place preference ratio
|
22.33 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Novelty place preference
|
20.99 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Time in novel zone, NPP test
|
17.06 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Total zone transitions, NPP test
|
106.24 |
1 |
1 |
3.2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Total locomotion distance, NPP test
|
47.64 |
1 |
1 |
3.2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Locomotion velocity, NPP test
|
51.12 |
1 |
1 |
3.2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Pavlov. Cond. change in total contacts
|
69.48 |
1 |
1 |
3.2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Pavlov. Cond. index score
|
14.07 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Pavlov. Cond. lever-magazine prob. diff.
|
14.82 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Bone: apparent density
|
12.01 |
6 |
0 |
0 |
1 |
2.48e-19 |
Fry
Hmgb1
Katnal1
LOC102549494
LOC102553253
Rxfp2
|
|
Bone surface
|
11.81 |
4 |
0 |
0 |
1 |
4.71e-16 |
Fry
LOC102549494
Rxfp2
Snapc2
|
|
Bone volume
|
11.59 |
4 |
0 |
0 |
0.99 |
1.09e-07 |
Cacul1
Hmgb1
LOC102549494
LOC102553253
|
|
Bone: connectivity density
|
13.7 |
6 |
1 |
3.2 |
1 |
2.96e-19 |
Fry
LOC102549494
LOC120095830
LOC120095871
Rxfp2
Snapc2
|
|
Bone: cortical area
|
14.22 |
11 |
0 |
0 |
1 |
1.70e-39 |
Cacul1
Ces2c
Dennd10
Emx2os
Fam204a
Grk5
LOC102550729
Prlhr
Rab11fip2
Sfxn4
Zfp950
|
|
Bone: cortical porosity
|
16.96 |
1 |
0 |
0 |
0 |
1.00e+00 |
LOC103694460
|
|
Bone: cortical thickness
|
11.55 |
6 |
0 |
0 |
-0.03 |
9.48e-01 |
Emx2os
Fam204a
LOC120100068
Prlhr
Rab11fip2
Zfp950
|
|
Bone: endosteal perimeter
|
25.18 |
1 |
1 |
3.2 |
0 |
1.00e+00 |
LOC103694460
|
|
Bone: final force
|
11.23 |
6 |
0 |
0 |
1 |
1.21e-12 |
Cacul1
Grk5
Prlhr
Rab11fip2
Sfxn4
Zfp950
|
|
Bone: final moment
|
11.73 |
6 |
0 |
0 |
1 |
3.58e-19 |
Cacul1
Grk5
Prlhr
Rab11fip2
Sfxn4
Zfp950
|
|
Bone: maximum diameter
|
23.35 |
1 |
1 |
3.2 |
0 |
1.00e+00 |
LOC103694460
|
|
Bone: maximum force
|
10.64 |
6 |
0 |
0 |
1 |
2.74e-10 |
Cacul1
Grk5
Prlhr
Rab11fip2
Sfxn4
Zfp950
|
|
Bone: maximum moment
|
11.11 |
6 |
0 |
0 |
1 |
3.98e-12 |
Cacul1
Grk5
Prlhr
Rab11fip2
Sfxn4
Zfp950
|
|
Bone: minimum diameter
|
9.28 |
2 |
0 |
0 |
0 |
1.00e+00 |
Cacul1
LOC103694460
|
|
Bone: periosteal estimation
|
9.62 |
9 |
0 |
0 |
1 |
2.80e-27 |
Cacul1
Dennd10
Emx2os
Fam204a
Grk5
Prlhr
Rab11fip2
Sfxn4
Zfp950
|
|
Bone: periosteal perimeter
|
12.04 |
9 |
0 |
0 |
0.99 |
3.21e-26 |
Cacul1
Ces2c
Dennd10
Grk5
LOC102550729
LOC103694460
Prlhr
Sfxn4
Zfp950
|
|
Bone: stiffness
|
13.41 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Bone: trabecular number
|
12.38 |
1 |
0 |
0 |
1 |
3.69e-08 |
LOC102549494
|
|
Bone: trabecular thickness
|
15.76 |
5 |
0 |
0 |
1 |
9.51e-11 |
Cacul1
Grk5
Prlhr
Sfxn4
Zfp950
|
|
Distance traveled before self-admin
|
50.41 |
1 |
1 |
3.2 |
0 |
1.00e+00 |
Zdhhc1
|
|
Delta time in open arm before/after self-admin
|
26.67 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zfand2b
|
|
Active lever presses in extinction session 6
|
26.41 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zfand2b
|
|
Delta ambulatory time before/after self-admin
|
16.57 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zdhhc1
|
|
Time in closed arm before self-admin
|
44.32 |
1 |
1 |
3.2 |
0 |
1.00e+00 |
Zfand2b
|
|
Time in open arm before self-admin
|
24.67 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zfand2b
|
|
Time to tail flick, vehicle, before self-admin
|
18.85 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zdhhc1
|
|
Time to tail flick, vehicle, after self-admin
|
56.6 |
2 |
2 |
6.5 |
0 |
1.00e+00 |
Klhdc8b
Zfand2b
|
|
Delta time to tail flick, vehicle, before/after SA
|
74.6 |
1 |
1 |
3.2 |
0 |
1.00e+00 |
Zfand2b
|
|
Ambulatory time before self-admin
|
29.88 |
2 |
1 |
3.2 |
0 |
1.00e+00 |
Zdhhc1
Zfand2b
|
|
Total heroin consumption
|
18.89 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Area under the delay curve
|
31.15 |
1 |
1 |
3.2 |
0 |
1.00e+00 |
Zfand2b
|
|
Delay disc. indifference point, 0s delay
|
28.7 |
1 |
1 |
3.2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Delay disc. indifference point, 16s delay
|
69.03 |
1 |
1 |
3.2 |
0 |
1.00e+00 |
Zfand2b
|
|
Delay disc. indifference point, 24s delay
|
27.43 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zfand2b
|
|
Delay disc. indifference point, 4s delay
|
41.82 |
1 |
1 |
3.2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Delay disc. indifference point, 8s delay
|
74.24 |
1 |
1 |
3.2 |
0 |
1.00e+00 |
Zfand2b
|
|
Delay discount exponential model param
|
16.14 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zfand2b
|
|
Fecal boli incidents, locomotor time 1
|
96.73 |
2 |
1 |
3.2 |
0 |
1.00e+00 |
Klhdc8b
Zfand2b
|
|
Time >=10cm from walls, locomotor time 1
|
79.04 |
1 |
1 |
3.2 |
0 |
1.00e+00 |
Zfand2b
|
|
Rest time, locomotor task time 1
|
22.55 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Distance moved, locomotor task time 1
|
31.01 |
1 |
1 |
3.2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Vertical activity count, locomotor time 1
|
73.36 |
2 |
2 |
6.5 |
0 |
1.00e+00 |
Klhdc8b
Zfand2b
|
|
Bouts of movement, locomotor time 2
|
21.88 |
2 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
Zfand2b
|
|
Total resting periods, locomotor time 2
|
28.25 |
2 |
1 |
3.2 |
0 |
1.00e+00 |
Klhdc8b
Zfand2b
|
|
Rest time, locomotor task time 2
|
28.62 |
3 |
2 |
6.5 |
0 |
1.00e+00 |
Klhdc8b
Zdhhc1
Zfand2b
|
|
Distance moved, locomotor task time 2
|
48.07 |
3 |
2 |
6.5 |
0 |
1.00e+00 |
Klhdc8b
Zdhhc1
Zfand2b
|
|
Vertical activity count, locomotor time 2
|
54.92 |
1 |
1 |
3.2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Weight adjusted by age
|
30.32 |
1 |
1 |
3.2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Progressive ratio
|
22.47 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zdhhc1
|
|
Food seeking constrained by brief footshock
|
14.98 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zfand2b
|
|
Run reversals in cocaine runway, females
|
36.45 |
1 |
1 |
3.2 |
0 |
1.00e+00 |
Zfand2b
|
|
Latency to leave start box in cocaine runway
|
34.23 |
1 |
1 |
3.2 |
0 |
1.00e+00 |
Zfand2b
|
|
Latency to leave start box in cocaine runway, F
|
31.9 |
1 |
1 |
3.2 |
0 |
1.00e+00 |
Zfand2b
|
|
Latency to leave start box in cocaine runway, M
|
42.07 |
1 |
1 |
3.2 |
0 |
1.00e+00 |
Zdhhc1
|
|
Cd content in liver
|
20.17 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Cu content in liver
|
43.87 |
1 |
1 |
3.2 |
0 |
1.00e+00 |
LOC103694460
|
|
Fe content in liver
|
33.68 |
2 |
2 |
6.5 |
0 |
1.00e+00 |
Klhdc8b
LOC103694460
|
|
K content in liver
|
46.75 |
1 |
1 |
3.2 |
0 |
1.00e+00 |
LOC103694460
|
|
Na content in liver
|
16.36 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Rb content in liver
|
66.6 |
2 |
2 |
6.5 |
0 |
1.00e+00 |
Klhdc8b
LOC103694460
|
|
Sr content in liver
|
22.32 |
2 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
LOC103694460
|