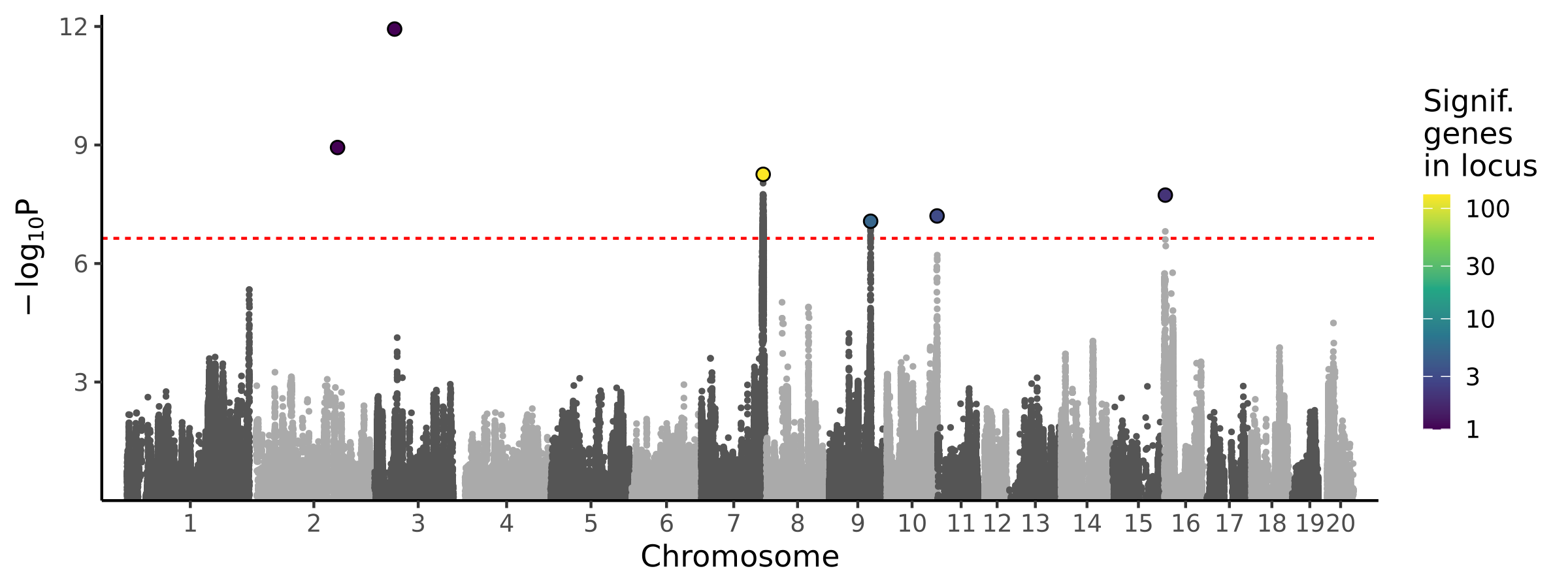
|
BMI with tail
|
4.63 |
34 |
0 |
0 |
1 |
1.76e-168 |
Aqp5
Asic1
Atf1
Bcdin3d
Cacnb3
Ccdc65
Cers5
Csrnp2
Dip2b
Dnajc22
Faim2
Kansl2
Kcnh3
Kmt2d
Larp4
Letmd1
Lima1
Lmbr1l
LOC100362820
LOC102546778
LOC108351553
LOC120093830
Mcrs1
Nckap5l
Prpf40b
Prph
Slc11a2
Smagp
Smarcd1
Spats2
Tmbim6
Tuba1a
Tuba1b
Tuba1c
|
|
Body weight
|
5.65 |
56 |
7 |
53.8 |
1 |
0.00e+00 |
Adcy6
Aqp5
Asic1
Atf1
B3gntl1
Bcdin3d
Bin2
C1ql4
Cacnb3
Ccdc65
Ccnt1
Cela1
Cers5
Csrnp2
Ddn
Ddx23
Dip2b
Dnajc22
Faim2
Fkbp11
Fmnl3
Kansl2
Kcnh3
Kmt2d
Larp4
Letmd1
Lima1
Lmbr1l
LOC100362820
LOC102546778
LOC102555574
LOC102556302
LOC103692978
LOC108351553
LOC108351554
LOC120093830
LOC120093831
LOC120093832
Mcrs1
Mettl7a
Nckap5l
Prkag1
Prpf40b
Prph
Rhebl1
Slc11a2
Slc4a8
Smagp
Smarcd1
Spats2
Tmbim6
Tmprss12
Tuba1a
Tuba1b
Tuba1c
Wnt10b
|
|
Left kidney weight
|
5.75 |
1 |
0 |
0 |
0 |
1.00e+00 |
B3gntl1
|
|
Length without tail
|
7.62 |
38 |
0 |
0 |
1 |
8.45e-232 |
Adcy6
Asic1
B3gntl1
Bcdin3d
C1ql4
Cacnb3
Ccdc65
Ccnt1
Ddn
Ddx23
Dnajc22
Faim2
Fkbp11
Fmnl3
Kansl2
Kcnh3
Kmt2d
Lmbr1l
LOC100362820
LOC102546778
LOC102555574
LOC102556302
LOC103692978
LOC120093830
LOC120093831
Ly75
Mcrs1
Nckap5l
Prkag1
Prpf40b
Prph
Rhebl1
Spats2
Tmbim6
Tuba1a
Tuba1b
Tuba1c
Wnt10b
|
|
Retroperitoneal fat weight
|
8.85 |
1 |
1 |
7.7 |
0 |
1.00e+00 |
Ly75
|
|
Extensor digitorum longus weight
|
4.33 |
43 |
0 |
0 |
1 |
6.06e-218 |
Aqp5
Asic1
Atf1
Bcdin3d
Bin2
C1ql4
Cacnb3
Ccdc65
Cers5
Csrnp2
Dip2b
Dnajc22
Faim2
Fmnl3
Kansl2
Kcnh3
Kmt2d
Larp4
Letmd1
Lima1
Lmbr1l
LOC100362820
LOC102546778
LOC102556302
LOC108351553
LOC108351554
LOC120093830
LOC120093832
Mcrs1
Mettl7a
Nckap5l
Prkag1
Prpf40b
Prph
Slc11a2
Slc4a8
Smagp
Smarcd1
Spats2
Tmbim6
Tuba1a
Tuba1b
Tuba1c
|
|
Tibialis anterior weight
|
4.45 |
1 |
0 |
0 |
1 |
6.20e-05 |
Slc11a2
|
|
Tibia length
|
6.87 |
54 |
1 |
7.7 |
0.99 |
5.13e-226 |
Adcy6
Aqp5
Asic1
Atf1
B3gntl1
Bcdin3d
Bin2
C1ql4
Cacnb3
Ccdc65
Ccnt1
Cela1
Csrnp2
Ddn
Ddx23
Dip2b
Dnajc22
Efhd1
Faim2
Fkbp11
Fmnl3
Kansl2
Kcnh3
Kmt2d
Larp4
Letmd1
Lmbr1l
LOC100362820
LOC102546778
LOC102555574
LOC102556302
LOC103692978
LOC108351553
LOC108351554
LOC120093830
LOC120093831
Mcrs1
Mettl7a
Nckap5l
Neu2
Prkag1
Prpf40b
Prph
Rhebl1
Slc11a2
Smagp
Smarcd1
Spats2
Tmbim6
Tmprss12
Tuba1a
Tuba1b
Tuba1c
Wnt10b
|
|
Food consumed during 24 hour testing period
|
11.76 |
1 |
0 |
0 |
0 |
1.00e+00 |
Ly75
|
|
Water consumed over 24 hours
|
19.18 |
1 |
0 |
0 |
0 |
1.00e+00 |
Ly75
|
|
Patch foraging total patch changes 18 sec
|
13.09 |
1 |
0 |
0 |
0 |
1.00e+00 |
Ly75
|
|
Reaction time mean minus median AUC
|
17.14 |
1 |
0 |
0 |
0 |
1.00e+00 |
Ly75
|
|
Reaction time mean AUC
|
10.73 |
1 |
0 |
0 |
0 |
1.00e+00 |
Ly75
|
|
Reaction time omissions
|
10.42 |
11 |
0 |
0 |
-1 |
1.95e-31 |
Ccdc65
Faim2
Kcnh3
LOC102546778
LOC120093830
Mcrs1
Prkag1
Spats2
Tmbim6
Tuba1a
Tuba1b
|
|
Reaction time premature initiation rate
|
20.51 |
1 |
1 |
7.7 |
0 |
1.00e+00 |
Ilf2
|
|
Reaction time premature initiations
|
12.93 |
1 |
0 |
0 |
0 |
1.00e+00 |
Ilf2
|
|
Std. dev. reaction times
|
14.29 |
1 |
0 |
0 |
0 |
1.00e+00 |
Ly75
|
|
Condit. Reinf. active minus inactive responses
|
28.62 |
1 |
1 |
7.7 |
0 |
1.00e+00 |
Ly75
|
|
Condit. Reinf. active responses
|
24.88 |
1 |
1 |
7.7 |
0 |
1.00e+00 |
Ly75
|
|
Condit. Reinf. lever reinforcers received
|
19.5 |
1 |
1 |
7.7 |
0 |
1.00e+00 |
Ly75
|
|
Pavlov. Cond. change in total contacts
|
11.21 |
1 |
0 |
0 |
0 |
1.00e+00 |
Ly75
|
|
Locomotion distance, session 3
|
14.48 |
1 |
0 |
0 |
0 |
1.00e+00 |
Ly75
|
|
Stereotopy head waving bouts, day 7
|
23.7 |
1 |
0 |
0 |
0 |
1.00e+00 |
Ly75
|
|
Stereotopy head waving duration, day 7
|
21.6 |
1 |
0 |
0 |
0 |
1.00e+00 |
Ly75
|
|
Locomotion velocity, session 8
|
12.45 |
1 |
0 |
0 |
0 |
1.00e+00 |
Ilf2
|
|
Degree of sensitization stereotypy
|
38.66 |
1 |
1 |
7.7 |
0 |
1.00e+00 |
Ly75
|
|
Condit. Reinf. active minus inactive responses
|
16.76 |
1 |
0 |
0 |
0 |
1.00e+00 |
Ly75
|
|
Condit. Reinf. active responses
|
13.63 |
1 |
0 |
0 |
0 |
1.00e+00 |
Ly75
|
|
Incentive salience index mean
|
15.42 |
1 |
0 |
0 |
0 |
1.00e+00 |
Ly75
|
|
Condit. Reinf. lever presses
|
10.84 |
1 |
0 |
0 |
0 |
1.00e+00 |
Ilf2
|
|
Total zone transitions, hab. session 2
|
19.52 |
1 |
0 |
0 |
0 |
1.00e+00 |
Ly75
|
|
Total locomotion distance, hab. session 2
|
24.97 |
1 |
1 |
7.7 |
0 |
1.00e+00 |
Ly75
|
|
Locomotion velocity, hab. session 2
|
19.99 |
1 |
1 |
7.7 |
0 |
1.00e+00 |
Ly75
|
|
Pavlov. Cond. lever latency
|
21.9 |
1 |
1 |
7.7 |
0 |
1.00e+00 |
Ilf2
|
|
Pavlov. Cond. magazine entry latency
|
28.91 |
1 |
1 |
7.7 |
0 |
1.00e+00 |
Ilf2
|
|
Pavlov. Cond. change in total contacts
|
18.74 |
1 |
0 |
0 |
0 |
1.00e+00 |
Ilf2
|
|
Pavlov. Cond. index score
|
34.08 |
1 |
1 |
7.7 |
0 |
1.00e+00 |
Ilf2
|
|
Pavlov. Cond. latency score
|
30.82 |
1 |
1 |
7.7 |
0 |
1.00e+00 |
Ilf2
|
|
Pavlov. Cond. lever contacts
|
37.24 |
1 |
1 |
7.7 |
0 |
1.00e+00 |
Ilf2
|
|
Pavlov. Cond. magazine entry number
|
42.75 |
1 |
1 |
7.7 |
0 |
1.00e+00 |
Ilf2
|
|
Pavlov. Cond. intertrial magazine entries
|
13.5 |
1 |
0 |
0 |
0 |
1.00e+00 |
Ilf2
|
|
Pavlov. Cond. lever-magazine prob. diff.
|
35.53 |
1 |
1 |
7.7 |
0 |
1.00e+00 |
Ilf2
|
|
Pavlov. Cond. response bias
|
41.38 |
1 |
1 |
7.7 |
0 |
1.00e+00 |
Ilf2
|
|
Bone: apparent density
|
10.76 |
44 |
1 |
7.7 |
1 |
7.38e-240 |
Adcy6
Aqp5
Asic1
Bcdin3d
C1ql4
Cacnb3
Ccdc65
Ccnt1
Csrnp2
Ddn
Ddx23
Dip2b
Dnajc22
Faim2
Fkbp11
Fmnl3
Kansl2
Kcnh3
Kmt2d
Lmbr1l
LOC100362820
LOC102546778
LOC102555574
LOC102556302
LOC103692978
LOC108351553
LOC108351554
LOC120093830
LOC120093831
Ly75
Mcrs1
Nckap5l
Prkag1
Prpf40b
Prph
Rhebl1
Slc11a2
Smarcd1
Spats2
Tmbim6
Tuba1a
Tuba1b
Tuba1c
Wnt10b
|
|
Bone surface
|
11.94 |
56 |
1 |
7.7 |
1 |
0.00e+00 |
Adcy6
Aqp5
Asic1
Atf1
Bcdin3d
Bin2
C1ql4
Cacnb3
Ccdc65
Ccnt1
Cela1
Cers5
Csrnp2
Ddn
Ddx23
Dip2b
Dnajc22
Faim2
Fkbp11
Fmnl3
Kansl2
Kcnh3
Kmt2d
Larp4
Letmd1
Lima1
Lmbr1l
LOC100362820
LOC102546778
LOC102555574
LOC102556302
LOC103692978
LOC108351553
LOC108351554
LOC120093830
LOC120093831
LOC120093832
Ly75
Mcrs1
Mettl7a
Nckap5l
Prkag1
Prpf40b
Prph
Rhebl1
Slc11a2
Slc4a8
Smagp
Smarcd1
Spats2
Tmbim6
Tmprss12
Tuba1a
Tuba1b
Tuba1c
Wnt10b
|
|
Bone volume
|
9.69 |
56 |
1 |
7.7 |
1 |
0.00e+00 |
Adcy6
Aqp5
Asic1
Atf1
Bcdin3d
Bin2
C1ql4
Cacnb3
Ccdc65
Ccnt1
Cela1
Cers5
Csrnp2
Ddn
Ddx23
Dip2b
Dnajc22
Faim2
Fkbp11
Fmnl3
Kansl2
Kcnh3
Kmt2d
Larp4
Letmd1
Lima1
Lmbr1l
LOC100362820
LOC102546778
LOC102555574
LOC102556302
LOC103692978
LOC108351553
LOC108351554
LOC120093830
LOC120093831
LOC120093832
Ly75
Mcrs1
Mettl7a
Nckap5l
Prkag1
Prpf40b
Prph
Rhebl1
Slc11a2
Slc4a8
Smagp
Smarcd1
Spats2
Tmbim6
Tmprss12
Tuba1a
Tuba1b
Tuba1c
Wnt10b
|
|
Bone: connectivity density
|
13.44 |
3 |
1 |
7.7 |
0 |
1.00e+00 |
Kansl2
Ly75
Tuba1b
|
|
Bone: cortical apparent density
|
18.66 |
1 |
0 |
0 |
0 |
1.00e+00 |
Ly75
|
|
Bone: cortical area
|
10.04 |
35 |
1 |
7.7 |
1 |
3.44e-107 |
Adcy6
B3gntl1
Bcdin3d
Cacnb3
Ccdc65
Ccnt1
Ddn
Ddx23
Dnajc22
Erc2
Faim2
Fkbp11
Fmnl3
Kansl2
Kcnh3
Kmt2d
Lmbr1l
LOC100362820
LOC102546778
LOC102555574
LOC102556302
LOC103692978
LOC120093830
LOC120093831
Ly75
Prkag1
Prpf40b
Prph
Rhebl1
Spats2
Tmbim6
Tuba1a
Tuba1b
Tuba1c
Wnt10b
|
|
Bone: cortical thickness
|
23.25 |
1 |
1 |
7.7 |
0 |
1.00e+00 |
B3gntl1
|
|
Bone: cortical thickness
|
12.83 |
1 |
0 |
0 |
0 |
1.00e+00 |
B3gntl1
|
|
Bone: cortical tissue density
|
17.88 |
1 |
0 |
0 |
0 |
1.00e+00 |
Ly75
|
|
Bone: elastic work
|
15.2 |
2 |
0 |
0 |
0.18 |
8.19e-01 |
B3gntl1
Ly75
|
|
Bone: endosteal estimation
|
8.11 |
58 |
0 |
0 |
1 |
0.00e+00 |
Adcy6
Aqp5
Asic1
Atf1
Bcdin3d
Bin2
C1ql4
Cacnb3
Ccdc65
Ccdc66
Ccnt1
Cela1
Cers5
Csrnp2
Ddn
Ddx23
Dip2b
Dnajc22
Erc2
Faim2
Fkbp11
Fmnl3
Kansl2
Kcnh3
Kmt2d
Larp4
Letmd1
Lima1
Lmbr1l
LOC100362820
LOC102546778
LOC102555574
LOC102556302
LOC103692978
LOC108351553
LOC108351554
LOC120093830
LOC120093831
LOC120093832
Ly75
Mcrs1
Mettl7a
Nckap5l
Prkag1
Prpf40b
Prph
Rhebl1
Slc11a2
Slc4a8
Smagp
Smarcd1
Spats2
Tmbim6
Tmprss12
Tuba1a
Tuba1b
Tuba1c
Wnt10b
|
|
Bone: endosteal perimeter
|
7.21 |
43 |
0 |
0 |
1 |
1.30e-250 |
Aqp5
Asic1
Atf1
Bcdin3d
Bin2
Cacnb3
Cela1
Cers5
Csrnp2
Dip2b
Dnajc22
Faim2
Fkbp11
Kansl2
Kcnh3
Kmt2d
Larp4
Letmd1
Lima1
Lmbr1l
LOC100362820
LOC102546778
LOC108351553
LOC108351554
LOC120093830
LOC120093832
Mcrs1
Mettl7a
Nckap5l
Prkag1
Prpf40b
Prph
Rhebl1
Slc11a2
Smagp
Smarcd1
Spats2
Tmbim6
Tmprss12
Tuba1a
Tuba1b
Tuba1c
Wnt10b
|
|
Bone: final force
|
11.37 |
62 |
3 |
23.1 |
1 |
0.00e+00 |
Adcy6
Aqp5
Asic1
Atf1
B3gntl1
Bcdin3d
Bin2
C1ql4
Cacnb3
Ccdc65
Ccdc66
Ccnt1
Cela1
Cers5
Csrnp2
Ddn
Ddx23
Dip2b
Dnajc22
Efhd1
Erc2
Faim2
Fkbp11
Fmnl3
Ilf2
Kansl2
Kcnh3
Kmt2d
Larp4
Letmd1
Lima1
Lmbr1l
LOC100362820
LOC102546778
LOC102555574
LOC102556302
LOC103692978
LOC108351553
LOC108351554
LOC120093830
LOC120093831
LOC120093832
Ly75
Mcrs1
Mettl7a
Nckap5l
Neu2
Prkag1
Prpf40b
Prph
Rhebl1
Slc11a2
Slc4a8
Smagp
Smarcd1
Spats2
Tmbim6
Tmprss12
Tuba1a
Tuba1b
Tuba1c
Wnt10b
|
|
Bone: final moment
|
12.57 |
62 |
4 |
30.8 |
1 |
0.00e+00 |
Adcy6
Aqp5
Asic1
Atf1
B3gntl1
Bcdin3d
Bin2
C1ql4
Cacnb3
Ccdc65
Ccdc66
Ccnt1
Cela1
Cers5
Csrnp2
Ddn
Ddx23
Dip2b
Dnajc22
Efhd1
Erc2
Faim2
Fkbp11
Fmnl3
Ilf2
Kansl2
Kcnh3
Kmt2d
Larp4
Letmd1
Lima1
Lmbr1l
LOC100362820
LOC102546778
LOC102555574
LOC102556302
LOC103692978
LOC108351553
LOC108351554
LOC120093830
LOC120093831
LOC120093832
Ly75
Mcrs1
Mettl7a
Nckap5l
Neu2
Prkag1
Prpf40b
Prph
Rhebl1
Slc11a2
Slc4a8
Smagp
Smarcd1
Spats2
Tmbim6
Tmprss12
Tuba1a
Tuba1b
Tuba1c
Wnt10b
|
|
Bone: marrow area
|
7.86 |
58 |
0 |
0 |
1 |
0.00e+00 |
Adcy6
Aqp5
Asic1
Atf1
Bcdin3d
Bin2
C1ql4
Cacnb3
Ccdc65
Ccdc66
Ccnt1
Cela1
Cers5
Csrnp2
Ddn
Ddx23
Dip2b
Dnajc22
Erc2
Faim2
Fkbp11
Fmnl3
Kansl2
Kcnh3
Kmt2d
Larp4
Letmd1
Lima1
Lmbr1l
LOC100362820
LOC102546778
LOC102555574
LOC102556302
LOC103692978
LOC108351553
LOC108351554
LOC120093830
LOC120093831
LOC120093832
Ly75
Mcrs1
Mettl7a
Nckap5l
Prkag1
Prpf40b
Prph
Rhebl1
Slc11a2
Slc4a8
Smagp
Smarcd1
Spats2
Tmbim6
Tmprss12
Tuba1a
Tuba1b
Tuba1c
Wnt10b
|
|
Bone: maximum force
|
15.27 |
62 |
28 |
215.4 |
1 |
0.00e+00 |
Adcy6
Aqp5
Asic1
Atf1
B3gntl1
Bcdin3d
Bin2
C1ql4
Cacnb3
Ccdc65
Ccdc66
Ccnt1
Cela1
Cers5
Csrnp2
Ddn
Ddx23
Dip2b
Dnajc22
Efhd1
Erc2
Faim2
Fkbp11
Fmnl3
Ilf2
Kansl2
Kcnh3
Kmt2d
Larp4
Letmd1
Lima1
Lmbr1l
LOC100362820
LOC102546778
LOC102555574
LOC102556302
LOC103692978
LOC108351553
LOC108351554
LOC120093830
LOC120093831
LOC120093832
Ly75
Mcrs1
Mettl7a
Nckap5l
Neu2
Prkag1
Prpf40b
Prph
Rhebl1
Slc11a2
Slc4a8
Smagp
Smarcd1
Spats2
Tmbim6
Tmprss12
Tuba1a
Tuba1b
Tuba1c
Wnt10b
|
|
Bone: minimum diameter
|
9.01 |
58 |
1 |
7.7 |
1 |
0.00e+00 |
Adcy6
Aqp5
Asic1
Atf1
Bcdin3d
Bin2
C1ql4
Cacnb3
Ccdc65
Ccdc66
Ccnt1
Cela1
Cers5
Csrnp2
Ddn
Ddx23
Dip2b
Dnajc22
Erc2
Faim2
Fkbp11
Fmnl3
Kansl2
Kcnh3
Kmt2d
Larp4
Letmd1
Lima1
Lmbr1l
LOC100362820
LOC102546778
LOC102555574
LOC102556302
LOC103692978
LOC108351553
LOC108351554
LOC120093830
LOC120093831
LOC120093832
Ly75
Mcrs1
Mettl7a
Nckap5l
Prkag1
Prpf40b
Prph
Rhebl1
Slc11a2
Slc4a8
Smagp
Smarcd1
Spats2
Tmbim6
Tmprss12
Tuba1a
Tuba1b
Tuba1c
Wnt10b
|
|
Bone: periosteal estimation
|
11.3 |
59 |
3 |
23.1 |
1 |
0.00e+00 |
Adcy6
Aqp5
Asic1
Atf1
Bcdin3d
Bin2
C1ql4
Cacnb3
Ccdc65
Ccdc66
Ccnt1
Cela1
Cers5
Csrnp2
Ddn
Ddx23
Dip2b
Dnajc22
Erc2
Faim2
Fkbp11
Fmnl3
Ilf2
Kansl2
Kcnh3
Kmt2d
Larp4
Letmd1
Lima1
Lmbr1l
LOC100362820
LOC102546778
LOC102555574
LOC102556302
LOC103692978
LOC108351553
LOC108351554
LOC120093830
LOC120093831
LOC120093832
Ly75
Mcrs1
Mettl7a
Nckap5l
Prkag1
Prpf40b
Prph
Rhebl1
Slc11a2
Slc4a8
Smagp
Smarcd1
Spats2
Tmbim6
Tmprss12
Tuba1a
Tuba1b
Tuba1c
Wnt10b
|
|
Bone: periosteal perimeter
|
10.07 |
56 |
0 |
0 |
1 |
0.00e+00 |
Adcy6
Aqp5
Asic1
Atf1
B3gntl1
Bcdin3d
Bin2
C1ql4
Cacnb3
Ccdc65
Ccnt1
Cela1
Cers5
Csrnp2
Ddn
Ddx23
Dip2b
Dnajc22
Faim2
Fkbp11
Fmnl3
Kansl2
Kcnh3
Kmt2d
Larp4
Letmd1
Lima1
Lmbr1l
LOC100362820
LOC102546778
LOC102555574
LOC102556302
LOC103692978
LOC108351553
LOC108351554
LOC120093830
LOC120093831
LOC120093832
Mcrs1
Mettl7a
Nckap5l
Prkag1
Prpf40b
Prph
Rhebl1
Slc11a2
Slc4a8
Smagp
Smarcd1
Spats2
Tmbim6
Tmprss12
Tuba1a
Tuba1b
Tuba1c
Wnt10b
|
|
Bone: post-yield work
|
11.27 |
3 |
0 |
0 |
0 |
1.00e+00 |
Dip2b
LOC100362820
LOC102546778
|
|
Bone: stiffness
|
18.26 |
59 |
55 |
423.1 |
1 |
0.00e+00 |
Adcy6
Aqp5
Asic1
Atf1
B3gntl1
Bcdin3d
Bin2
C1ql4
Cacnb3
Ccdc65
Ccdc66
Ccnt1
Cela1
Cers5
Csrnp2
Ddn
Ddx23
Dip2b
Dnajc22
Erc2
Faim2
Fkbp11
Fmnl3
Ilf2
Kansl2
Kcnh3
Kmt2d
Larp4
Letmd1
Lima1
Lmbr1l
LOC100362820
LOC102546778
LOC102555574
LOC102556302
LOC103692978
LOC108351553
LOC108351554
LOC120093830
LOC120093831
LOC120093832
Mcrs1
Mettl7a
Nckap5l
Prkag1
Prpf40b
Prph
Rhebl1
Slc11a2
Slc4a8
Smagp
Smarcd1
Spats2
Tmbim6
Tmprss12
Tuba1a
Tuba1b
Tuba1c
Wnt10b
|
|
Bone: tissue strength
|
16.93 |
1 |
0 |
0 |
1 |
9.83e-09 |
Efhd1
|
|
Bone: trabecular number
|
22.37 |
2 |
1 |
7.7 |
0 |
1.00e+00 |
Ly75
Tuba1b
|
|
Bone: trabecular spacing
|
60.24 |
1 |
1 |
7.7 |
0 |
1.00e+00 |
Ly75
|
|
Bone: trabecular thickness
|
47.42 |
1 |
1 |
7.7 |
0 |
1.00e+00 |
Ly75
|
|
Distance moved, locomotor task time 1
|
13.8 |
1 |
0 |
0 |
0 |
1.00e+00 |
Ly75
|
|
Weight adjusted by age
|
20.87 |
1 |
0 |
0 |
0 |
1.00e+00 |
Ly75
|
|
Locomotion in novel chamber
|
34.01 |
1 |
1 |
7.7 |
0 |
1.00e+00 |
Ly75
|
|
Progressive ratio
|
12.89 |
1 |
0 |
0 |
0 |
1.00e+00 |
Ilf2
|
|
Food seeking constrained by brief footshock
|
13.79 |
1 |
0 |
0 |
0 |
1.00e+00 |
Ly75
|
|
Run reversals in cocaine runway, females
|
21.72 |
1 |
0 |
0 |
0 |
1.00e+00 |
Ly75
|
|
Latency to leave start box in cocaine runway, M
|
14.76 |
1 |
0 |
0 |
0 |
1.00e+00 |
Ilf2
|
|
Cd content in liver
|
13.38 |
1 |
0 |
0 |
0 |
1.00e+00 |
Ilf2
|
|
Mg content in liver
|
13.79 |
1 |
0 |
0 |
0 |
1.00e+00 |
Ly75
|
|
Mn content in liver
|
32.26 |
1 |
1 |
7.7 |
0 |
1.00e+00 |
Ly75
|
|
Zn content in liver
|
12.81 |
1 |
0 |
0 |
0 |
1.00e+00 |
Ly75
|