Pavlov. Cond. index score
Pavlovian Conditioned Approach, Sign/goal-tracking, average of LatencyScore, ProbDiff, and ResponseBias on days 4 and 5 of testing
Tags: Behavior · Pavlovian conditioned approach · Conditioned reinforcement
Project: p50_paul_meyer_2014
2 loci · 2 genes with independent associations · 2 total associated genes
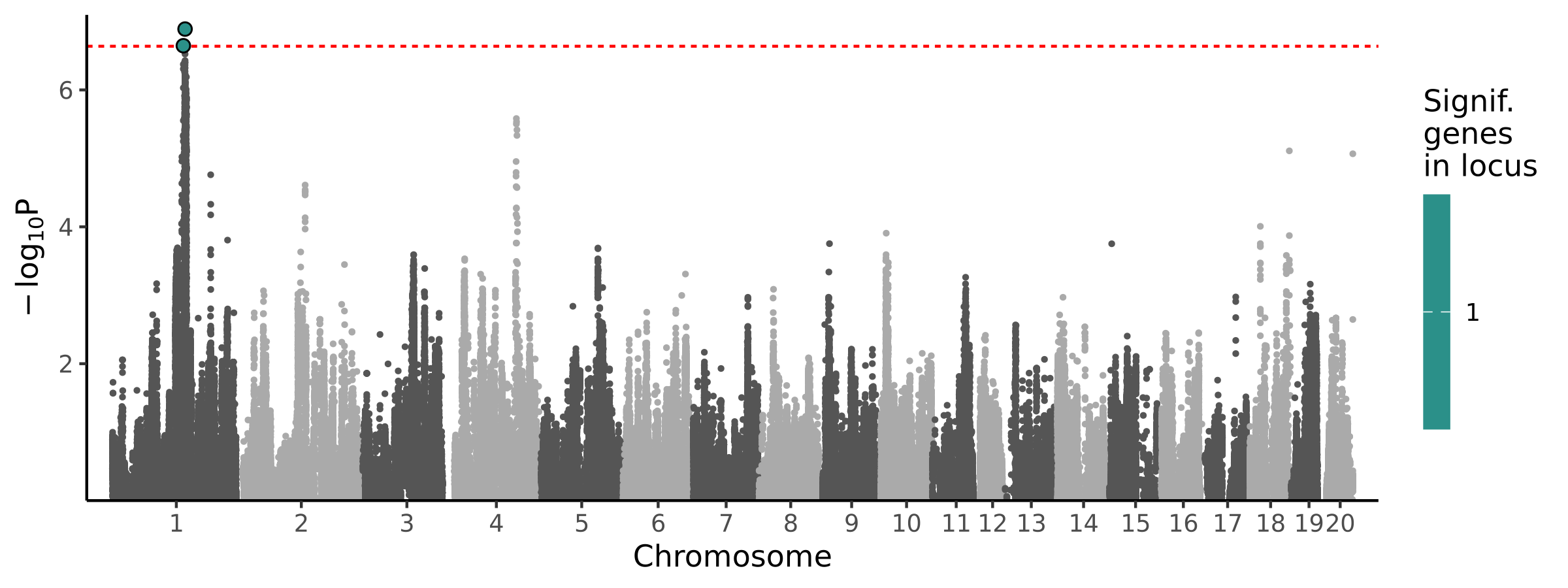
Significant Loci
| # | Chr | Start pos | End pos | # assoc genes | # joint models | Best TWAS P | Best GWAS P | Cond GWAS P | Joint genes |
|---|---|---|---|---|---|---|---|---|---|
| 1 | chr1 | 149199615 | 150594931 | 1 | 1 | 2.26e-07 | 1.49e-07 | 1.42e-01 | Tenm4 |
| 2 | chr1 | 152785136 | 154174144 | 1 | 1 | 1.29e-07 | 9.70e-07 | 5.99e-01 | Dgat2 |
Pleiotropic Associations
Associations by panel
| Tissue | RNA modality | # hits | % hits/tests | Avg chisq |
|---|---|---|---|---|
| Adipose | alternative polyA | 0 | 0 | 1.29 |
| Adipose | alternative TSS | 0 | 0 | 1.35 |
| Adipose | gene expression | 0 | 0 | 1.28 |
| Adipose | isoform ratio | 0 | 0 | 1.29 |
| Adipose | intron excision ratio | 0 | 0 | 1.12 |
| Adipose | mRNA stability | 0 | 0 | 1.31 |
| BLA | alternative polyA | 0 | 0 | 1.34 |
| BLA | alternative TSS | 0 | 0 | 1.29 |
| BLA | gene expression | 0 | 0 | 1.3 |
| BLA | isoform ratio | 0 | 0 | 1.33 |
| BLA | intron excision ratio | 0 | 0 | 1.25 |
| BLA | mRNA stability | 0 | 0 | 1.31 |
| Brain | alternative polyA | 0 | 0 | 1.3 |
| Brain | alternative TSS | 0 | 0 | 1.27 |
| Brain | gene expression | 0 | 0 | 1.28 |
| Brain | isoform ratio | 0 | 0 | 1.3 |
| Brain | intron excision ratio | 0 | 0 | 1.24 |
| Brain | mRNA stability | 0 | 0 | 1.27 |
| Eye | alternative polyA | 0 | 0 | 1.43 |
| Eye | alternative TSS | 0 | 0 | 1.27 |
| Eye | gene expression | 0 | 0 | 1.37 |
| Eye | isoform ratio | 0 | 0 | 1.25 |
| Eye | intron excision ratio | 0 | 0 | 1.29 |
| Eye | mRNA stability | 0 | 0 | 1.38 |
| IL | alternative polyA | 0 | 0 | 1.25 |
| IL | alternative TSS | 0 | 0 | 1.1 |
| IL | gene expression | 0 | 0 | 1.3 |
| IL | isoform ratio | 0 | 0 | 1.31 |
| IL | intron excision ratio | 0 | 0 | 1.24 |
| IL | mRNA stability | 0 | 0 | 1.29 |
| LHb | alternative polyA | 0 | 0 | 1.36 |
| LHb | alternative TSS | 0 | 0 | 1.43 |
| LHb | gene expression | 0 | 0 | 1.27 |
| LHb | isoform ratio | 0 | 0 | 1.3 |
| LHb | intron excision ratio | 0 | 0 | 1.22 |
| LHb | mRNA stability | 0 | 0 | 1.26 |
| Liver | alternative polyA | 0 | 0 | 1.25 |
| Liver | alternative TSS | 0 | 0 | 1.28 |
| Liver | gene expression | 1 | 0 | 1.32 |
| Liver | isoform ratio | 0 | 0 | 1.29 |
| Liver | intron excision ratio | 0 | 0 | 1.3 |
| Liver | mRNA stability | 0 | 0 | 1.32 |
| NAcc | alternative polyA | 0 | 0 | 1.36 |
| NAcc | alternative TSS | 0 | 0 | 1.26 |
| NAcc | gene expression | 1 | 0 | 1.28 |
| NAcc | isoform ratio | 0 | 0 | 1.3 |
| NAcc | intron excision ratio | 0 | 0 | 1.22 |
| NAcc | mRNA stability | 0 | 0 | 1.28 |
| OFC | alternative polyA | 0 | 0 | 1.38 |
| OFC | alternative TSS | 0 | 0 | 1.26 |
| OFC | gene expression | 0 | 0 | 1.27 |
| OFC | isoform ratio | 0 | 0 | 1.34 |
| OFC | intron excision ratio | 0 | 0 | 1.23 |
| OFC | mRNA stability | 0 | 0 | 1.29 |
| PL | alternative polyA | 0 | 0 | 1.43 |
| PL | alternative TSS | 0 | 0 | 1.33 |
| PL | gene expression | 0 | 0 | 1.29 |
| PL | isoform ratio | 0 | 0 | 1.28 |
| PL | intron excision ratio | 0 | 0 | 1.23 |
| PL | mRNA stability | 0 | 0 | 1.29 |
| pVTA | alternative polyA | 0 | 0 | 1.37 |
| pVTA | alternative TSS | 0 | 0 | 1.29 |
| pVTA | gene expression | 0 | 0 | 1.3 |
| pVTA | isoform ratio | 0 | 0 | 1.27 |
| pVTA | intron excision ratio | 0 | 0 | 1.21 |
| pVTA | mRNA stability | 0 | 0 | 1.29 |
| RMTg | alternative polyA | 0 | 0 | 1.15 |
| RMTg | alternative TSS | 0 | 0 | 1.14 |
| RMTg | gene expression | 0 | 0 | 1.31 |
| RMTg | isoform ratio | 0 | 0 | 1.11 |
| RMTg | intron excision ratio | 0 | 0 | 1.23 |
| RMTg | mRNA stability | 0 | 0 | 1.28 |