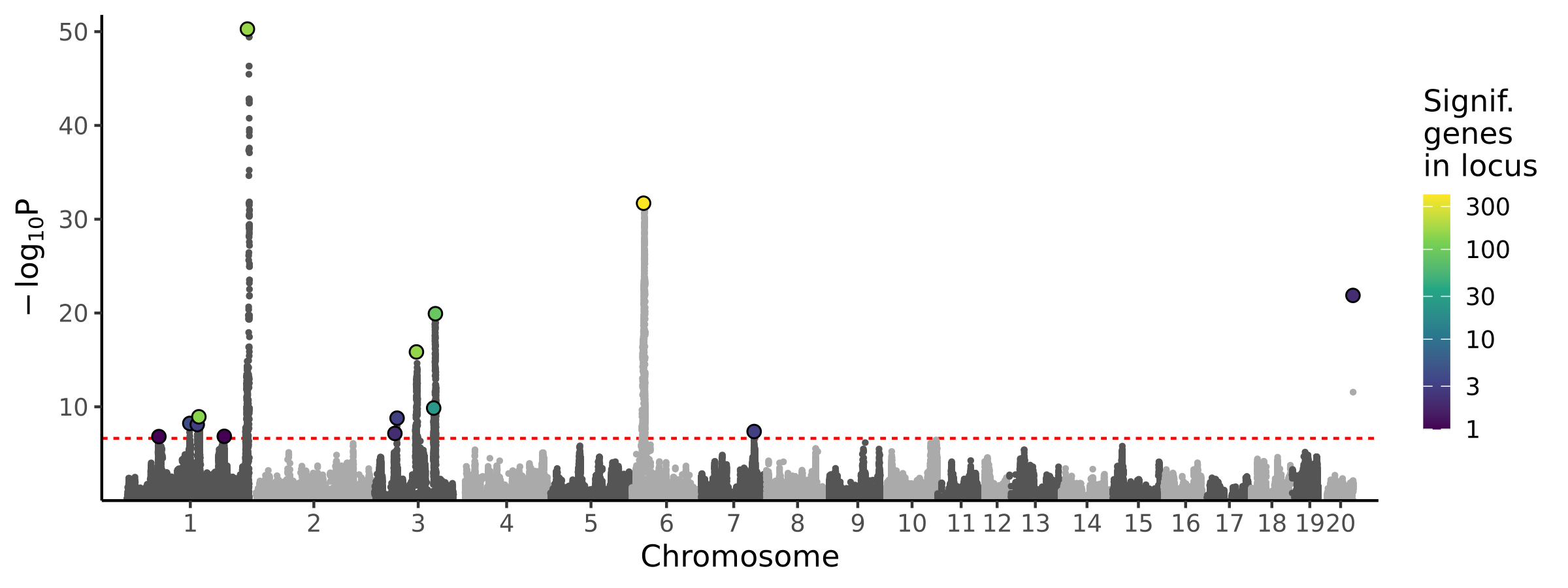
|
BMI with tail
|
12.19 |
66 |
55 |
112.2 |
1 |
0.00e+00 |
Ablim1
Acsl5
Adrb1
Afap1l2
Atrnl1
Cacul1
Casp7
Ccdc186
Cd99
Ces2c
Dclre1a
Dennd10
Dstn
Dtd1
Dzank1
Emx2os
Eno4
Fam204a
Fhip2a
Flrt3
Gfra1
Gpam
Grk5
Gucy2g
Habp2
Hspa12a
Kif16b
LOC102547573
LOC102548513
LOC102550304
LOC102550729
LOC102551125
LOC103694460
LOC120100064
LOC120100065
LOC120100068
LOC120101717
LOC120101719
Macrod2
Nanos1
Ndufaf5
Nhlrc2
Nrap
Otor
Pcsk2
Plekhs1
Pnlip
Polr3f
Prdx3
Prlhr
Rab11fip2
Rbbp9
Rrbp1
Scp2d1
Sec23b
Sfxn4
Smim26
Snrpb2
Tasp1
Tcf7l2
Tectb
Trub1
Vti1a
Zdhhc6
Zfp133
Zfp950
|
|
BMI without tail
|
14.73 |
37 |
15 |
30.6 |
0.99 |
9.22e-173 |
Aamdc
Alg8
Aqp11
B3gnt6
Cacul1
Ces2c
Clns1a
Dennd10
Dstn
Emx2os
Fam204a
Gab2
Grk5
Hspa12a
Ints4
Kif16b
LOC102550562
LOC102550729
LOC120100068
LOC120101717
LOC120101719
Myo7a
Nanos1
Nars2
Ndufc2
Otor
Pcsk2
Prdx3
Prlhr
Rab11fip2
Rrbp1
Sec23b
Sfxn4
Snrpb2
Tenm4
Usp35
Zfp950
|
|
Body weight
|
12.1 |
85 |
70 |
142.9 |
1 |
0.00e+00 |
Aamdc
Ablim1
Acsl5
Add3
Adrb1
Afap1l2
Alg8
Atrnl1
Bfsp1
Cacul1
Casp7
Ccdc186
Cd99
Ces2c
Dclre1a
Dennd10
Dstn
Dtd1
Dzank1
Efr3b
Emx2os
Eno4
Esf1
Fam204a
Fhip2a
Flrt3
Gab2
Gfra1
Gpam
Grk5
Gucy2g
Habp2
Hspa12a
Ism1
Kif16b
LOC102547573
LOC102548513
LOC102550304
LOC102550729
LOC102551125
LOC102551352
LOC103694460
LOC120100058
LOC120100064
LOC120100065
LOC120100066
LOC120100068
LOC120101717
LOC120101719
Macrod2
Mxi1
Nanos1
Nars2
Ndufaf5
Nhlrc2
Nrap
Otor
Pcsk2
Plekhs1
Pnlip
Polr3f
Prdx3
Prlhr
Rab11fip2
Rbbp9
Rrbp1
Scp2d1
Sec23b
Sfxn4
Shoc2
Smc3
Smim26
Snrpb2
Sptlc3
Tasp1
Tcf7l2
Tectb
Tenm4
Trub1
Usp35
Vti1a
Xpnpep1
Zdhhc6
Zfp133
Zfp950
|
|
Epididymis fat weight
|
15.98 |
130 |
110 |
224.5 |
0.99 |
0.00e+00 |
Abhd1
Ablim1
Acsl5
Adcy3
Adrb1
Afap1l2
Agbl5
Alk
Asxl2
Atraid
Atrnl1
Babam2
Cacul1
Cad
Casp7
Ccdc186
Cenpo
Ces2c
Cgref1
Clip4
Dclre1a
Dennd10
Dnajc27
Dnmt3a
Dpysl5
Drc1
Dstn
Dtnb
Efr3b
Eif2b4
Emx2os
Eno4
Fam204a
Fam228b
Fhip2a
Fkbp1b
Flrt3
Fndc4
Fosl2
Garem2
Gckr
Gfra1
Gpam
Gpn1
Grk5
Gtf3c2
Gucy2g
Habp2
Hadha
Hadhb
Hspa12a
Ift172
Ism1
Itsn2
Kcnk3
Kif16b
Kif3c
Klhl29
Krtcap3
Lbh
LOC102547573
LOC102548513
LOC102548914
LOC102550729
LOC102551125
LOC103692581
LOC120100064
LOC120100065
LOC120100066
LOC120100068
LOC120101717
LOC120101719
LOC120103492
LOC120103499
LOC120103502
Macrod2
Mpv17
Mrpl33
Mxi1
Nanos1
Ncoa1
Ndufaf5
Nhlrc2
Nrap
Nrbp1
Ost4
Otof
Otor
Pcare
Pcsk2
Plb1
Plekhs1
Pnlip
Ppm1g
Ppp1cb
Prdx3
Preb
Prlhr
Rab10
Rab11fip2
Rbbp9
Rbks
RGD1565766
Rrbp1
Sec23b
Sf3b6
Sfxn4
Slc30a3
Slc35f6
Slc4a1ap
Slc5a6
Snrpb2
Snx17
Spdya
Sptlc3
Supt7l
Tasp1
Tcf7l2
Tectb
Togaram2
Trim54
Trub1
Ubxn2a
Vti1a
Wdcp
Ypel5
Zdhhc6
Zfp512
Zfp513
Zfp950
|
|
Heart weight
|
14.17 |
1 |
0 |
0 |
0 |
1.00e+00 |
Rbks
|
|
Left kidney weight
|
10.9 |
27 |
18 |
36.7 |
0.97 |
4.30e-70 |
Acsl5
Cacul1
Casp7
Cbx7
Cd99
Ces2c
Dennd10
Emx2os
Fam204a
Gpam
Grk5
Gucy2g
Hspa12a
LOC102550729
LOC103694460
LOC120100064
LOC120100068
Mgat3
Nanos1
Prdx3
Prlhr
Rab11fip2
Sfxn4
Syngr1
Tcf7l2
Vti1a
Zfp950
|
|
Right kidney weight
|
10.05 |
40 |
24 |
49 |
0.97 |
1.25e-85 |
Ablim1
Acsl5
Adrb1
Afap1l2
Atrnl1
Cacul1
Casp7
Cbx7
Ces2c
Dclre1a
Dennd10
Emx2os
Fam204a
Fhip2a
Gpam
Grk5
Gucy2g
Habp2
Hspa12a
LOC102547573
LOC102550729
LOC103694460
LOC120100064
LOC120100065
LOC120100066
LOC120100068
Mgat3
Nanos1
Nhlrc2
Nrap
Plekhs1
Prdx3
Prlhr
Rab11fip2
Sfxn4
Syngr1
Tcf7l2
Trub1
Vti1a
Zfp950
|
|
Tail length
|
6.44 |
2 |
0 |
0 |
0 |
1.00e+00 |
Aamdc
Gab2
|
|
Length with tail
|
5.32 |
4 |
0 |
0 |
0.99 |
4.32e-08 |
Aamdc
Kif16b
Otor
Snrpb2
|
|
Length without tail
|
8.78 |
4 |
0 |
0 |
0.81 |
1.93e-01 |
Cacul1
Grk5
Ly75
Prlhr
|
|
Liver weight, right
|
13.85 |
18 |
6 |
12.2 |
0.98 |
7.11e-29 |
Cacul1
Casp7
Cd99
Clip4
Dennd10
Fam204a
Grk5
Habp2
LOC103694460
Nanos1
Plekhs1
Prlhr
Rab11fip2
Sfxn4
Spdya
Togaram2
Vti1a
Zfp950
|
|
Parametrial fat weight
|
14.1 |
87 |
47 |
95.9 |
0.98 |
0.00e+00 |
Ablim1
Acsl5
Adcy3
Afap1l2
Agbl5
Asxl2
Atrnl1
Bfsp1
Cacul1
Casp7
Ccdc186
Cenpo
Ces2c
Dclre1a
Dennd10
Dnajc27
Dnmt3a
Drc1
Dstn
Dtd1
Dtnb
Dzank1
Efr3b
Emx2os
Eno4
Fam204a
Fam228b
Fhip2a
Fkbp1b
Garem2
Gfra1
Gpam
Grk5
Gucy2g
Habp2
Hadha
Hadhb
Hspa12a
Itsn2
Kcnk3
Kif16b
Kif3c
Klhl29
LOC102547573
LOC102550304
LOC102550729
LOC102551125
LOC103692581
LOC120100065
LOC120100068
LOC120101717
LOC120101719
LOC120103492
LOC120103499
LOC120103502
Nanos1
Ncoa1
Nhlrc2
Nrap
Otof
Otor
Pcsk2
Plekhs1
Pnlip
Polr3f
Prdx3
Prlhr
Rab10
Rab11fip2
Rbbp9
RGD1565766
Rrbp1
Scp2d1
Sec23b
Sf3b6
Sfxn4
Slc35f6
Slc5a6
Smim26
Snrpb2
Tcf7l2
Trub1
Ubxn2a
Wdcp
Zdhhc6
Zfp133
Zfp950
|
|
Intraocular pressure
|
24.4 |
1 |
1 |
2 |
0 |
1.00e+00 |
LOC103694460
|
|
Extensor digitorum longus weight
|
4.84 |
32 |
0 |
0 |
-0.99 |
2.32e-315 |
Abhd1
Alk
Atraid
Babam2
Clip4
Eif2b4
Fndc4
Fosl2
Gpn1
Gtf3c2
Ift172
Krtcap3
LOC102548914
LOC103694460
Mpv17
Mrpl33
Nrbp1
Pcare
Plb1
Ppp1cb
Preb
Rbks
Slc4a1ap
Slc5a6
Snx17
Spdya
Supt7l
Togaram2
Trim54
Ypel5
Zfp512
Zfp513
|
|
Soleus weight
|
6.55 |
13 |
0 |
0 |
0.38 |
2.15e-02 |
Ablim1
Acsl5
Alk
Atrnl1
Fosl2
Gpam
Grk5
Gucy2g
LOC120100068
Tectb
Togaram2
Trub1
Vti1a
|
|
Tibialis anterior weight
|
11.7 |
1 |
1 |
2 |
0 |
1.00e+00 |
LOC103694460
|
|
Tibia length
|
7.61 |
12 |
0 |
0 |
0.98 |
1.40e-30 |
Esf1
Flrt3
Ism1
Kif16b
LOC102548513
LOC120101717
Macrod2
Ndufaf5
Otor
Snrpb2
Sptlc3
Tasp1
|
|
Water consumed over 24 hours
|
19.18 |
1 |
0 |
0 |
0 |
1.00e+00 |
Ly75
|
|
Patch foraging indifference point 0 sec
|
12.96 |
1 |
0 |
0 |
0 |
1.00e+00 |
LOC103694460
|
|
Patch foraging total patch changes 0 sec
|
12.64 |
1 |
0 |
0 |
0 |
1.00e+00 |
LOC103694460
|
|
Patch foraging total patch changes 6 sec
|
13.11 |
1 |
0 |
0 |
0 |
1.00e+00 |
Gfra1
|
|
Patch foraging water rate 18 sec
|
13.57 |
3 |
0 |
0 |
-1 |
6.40e-05 |
Dtd1
Scp2d1
Smim26
|
|
Reaction time mean minus median AUC
|
17.14 |
1 |
0 |
0 |
0 |
1.00e+00 |
Ly75
|
|
Std. dev. reaction times
|
14.29 |
1 |
0 |
0 |
0 |
1.00e+00 |
Ly75
|
|
Condit. Reinf. active minus inactive responses
|
28.62 |
1 |
1 |
2 |
0 |
1.00e+00 |
Ly75
|
|
Condit. Reinf. active responses
|
24.88 |
1 |
1 |
2 |
0 |
1.00e+00 |
Ly75
|
|
Condit. Reinf. lever presses
|
15.07 |
9 |
0 |
0 |
-1 |
3.72e-20 |
B3gnt6
Capn5
Gdpd4
LOC102550562
Lrrc32
Myo7a
Thap12
Tsku
Wnt11
|
|
Condit. Reinf. lever reinforcers received
|
19.5 |
1 |
1 |
2 |
0 |
1.00e+00 |
Ly75
|
|
Pavlov. Cond. lever latency
|
15.82 |
11 |
0 |
0 |
1 |
1.63e-22 |
B3gnt6
Capn5
Fam168a
Gdpd4
LOC100912071
LOC102550562
Lrrc32
Myo7a
Thap12
Tsku
Wnt11
|
|
Pavlov. Cond. change in total contacts
|
13.46 |
1 |
0 |
0 |
0 |
1.00e+00 |
Thap12
|
|
Pavlov. Cond. index score
|
14.76 |
4 |
0 |
0 |
-1 |
4.02e-04 |
LOC102550562
LOC103694460
Lrrc32
Thap12
|
|
Pavlov. Cond. latency score
|
14.82 |
1 |
0 |
0 |
0 |
1.00e+00 |
Thap12
|
|
Pavlov. Cond. lever contacts
|
16.16 |
11 |
0 |
0 |
-1 |
7.33e-23 |
B3gnt6
Capn5
Fam168a
Gdpd4
LOC100912071
LOC102550562
Lrrc32
Myo7a
Thap12
Tsku
Wnt11
|
|
Pavlov. Cond. magazine entry number
|
20.26 |
1 |
0 |
0 |
0 |
1.00e+00 |
LOC103694460
|
|
Pavlov. Cond. response bias
|
15.81 |
14 |
0 |
0 |
-1 |
1.01e-27 |
B3gnt6
Capn5
Coa4
Dnajb13
Fam168a
Gdpd4
LOC100912071
LOC102550562
Lrrc32
Map6
Myo7a
Thap12
Tsku
Wnt11
|
|
Conditioned reinforcement - inactives
|
17.21 |
6 |
0 |
0 |
1 |
1.04e-15 |
Ccdc73
Cstf3
Eif3m
Rcn1
Tcp11l1
Wt1
|
|
Intermitt. access day 15 inactive lever presses
|
20.17 |
2 |
0 |
0 |
0.81 |
1.94e-01 |
Gpn1
Gtf3c2
|
|
Progressive ratio test 1 inactive lever presses
|
19.16 |
1 |
0 |
0 |
0 |
1.00e+00 |
Gtf3c2
|
|
Stereotopy head waving bouts, day 7
|
23.7 |
1 |
0 |
0 |
0 |
1.00e+00 |
Ly75
|
|
Stereotopy head waving duration, day 7
|
21.6 |
1 |
0 |
0 |
0 |
1.00e+00 |
Ly75
|
|
Degree of sensitization stereotypy
|
38.66 |
1 |
1 |
2 |
0 |
1.00e+00 |
Ly75
|
|
Condit. Reinf. active minus inactive responses
|
16.76 |
1 |
0 |
0 |
0 |
1.00e+00 |
Ly75
|
|
Incentive salience index mean
|
15.42 |
1 |
0 |
0 |
0 |
1.00e+00 |
Ly75
|
|
Total zone transitions, hab. session 2
|
19.52 |
1 |
0 |
0 |
0 |
1.00e+00 |
Ly75
|
|
Total locomotion distance, hab. session 2
|
24.97 |
1 |
1 |
2 |
0 |
1.00e+00 |
Ly75
|
|
Locomotion velocity, hab. session 2
|
19.99 |
1 |
1 |
2 |
0 |
1.00e+00 |
Ly75
|
|
Bone: apparent density
|
27.67 |
1 |
1 |
2 |
0 |
1.00e+00 |
Ly75
|
|
Bone surface
|
47.59 |
1 |
1 |
2 |
0 |
1.00e+00 |
Ly75
|
|
Bone volume
|
43.15 |
1 |
1 |
2 |
0 |
1.00e+00 |
Ly75
|
|
Bone: connectivity density
|
21.24 |
1 |
1 |
2 |
0 |
1.00e+00 |
Ly75
|
|
Bone: cortical apparent density
|
18.66 |
1 |
0 |
0 |
0 |
1.00e+00 |
Ly75
|
|
Bone: cortical area
|
13.69 |
26 |
0 |
0 |
0.96 |
8.11e-47 |
Ablim1
Acsl5
Afap1l2
Cacul1
Casp7
Ccdc186
Ces2c
Dclre1a
Dennd10
Emx2os
Fam204a
Fhip2a
Gfra1
Grk5
Habp2
LOC102547573
LOC102550729
LOC120100065
LOC120100068
Ly75
Nhlrc2
Plekhs1
Prlhr
Rab11fip2
Sfxn4
Zfp950
|
|
Bone: cortical porosity
|
16.96 |
1 |
0 |
0 |
0 |
1.00e+00 |
LOC103694460
|
|
Bone: cortical thickness
|
14.35 |
9 |
3 |
6.1 |
0.92 |
5.13e-10 |
Emx2os
Esf1
LOC120100068
Macrod2
Ndufaf5
Prlhr
Rab11fip2
Sptlc3
Tasp1
|
|
Bone: cortical tissue density
|
17.88 |
1 |
0 |
0 |
0 |
1.00e+00 |
Ly75
|
|
Bone: elastic work
|
15.82 |
1 |
0 |
0 |
0 |
1.00e+00 |
Ly75
|
|
Bone: endosteal estimation
|
9.05 |
1 |
0 |
0 |
0 |
1.00e+00 |
Ly75
|
|
Bone: endosteal perimeter
|
18.12 |
2 |
1 |
2 |
0 |
1.00e+00 |
Cd99
LOC103694460
|
|
Bone: final force
|
15 |
5 |
1 |
2 |
0.73 |
6.31e-02 |
Cacul1
Grk5
Ly75
Prlhr
Zfp950
|
|
Bone: final moment
|
15.17 |
6 |
1 |
2 |
0.79 |
2.46e-03 |
Cacul1
Grk5
Ly75
Prlhr
Sfxn4
Zfp950
|
|
Bone: marrow area
|
9.02 |
1 |
0 |
0 |
0 |
1.00e+00 |
Ly75
|
|
Bone: maximum diameter
|
11.29 |
17 |
1 |
2 |
0.99 |
2.83e-29 |
Ablim1
Acsl5
Afap1l2
Cacul1
Casp7
Ccdc186
Cd99
Dclre1a
Fhip2a
Gfra1
Habp2
LOC102547573
LOC103694460
LOC120100065
Nhlrc2
Nrap
Plekhs1
|
|
Bone: maximum force
|
12.05 |
6 |
1 |
2 |
0.79 |
1.84e-02 |
Cacul1
Grk5
Ly75
Prlhr
Sfxn4
Zfp950
|
|
Bone: maximum moment
|
13.5 |
6 |
1 |
2 |
0.76 |
1.82e-02 |
Cacul1
Grk5
Ly75
Prlhr
Sfxn4
Zfp950
|
|
Bone: periosteal estimation
|
10.36 |
11 |
1 |
2 |
0.93 |
1.71e-09 |
Acsl5
Cacul1
Dennd10
Fam204a
Grk5
Ly75
Plekhs1
Prlhr
Rab11fip2
Sfxn4
Zfp950
|
|
Bone: periosteal perimeter
|
13.25 |
27 |
3 |
6.1 |
0.95 |
5.40e-79 |
Ablim1
Acsl5
Afap1l2
Atrnl1
Cacul1
Casp7
Ccdc186
Ces2c
Dclre1a
Dennd10
Fam204a
Fhip2a
Gfra1
Grk5
Habp2
LOC102547573
LOC102550729
LOC102551125
LOC103694460
LOC120100065
Nhlrc2
Nrap
Plekhs1
Rab11fip2
Sfxn4
Trub1
Zfp950
|
|
Bone: trabecular number
|
35.45 |
1 |
1 |
2 |
0 |
1.00e+00 |
Ly75
|
|
Bone: trabecular spacing
|
60.24 |
1 |
1 |
2 |
0 |
1.00e+00 |
Ly75
|
|
Bone: trabecular thickness
|
19.05 |
4 |
1 |
2 |
0.8 |
5.06e-03 |
Cacul1
Ly75
Prlhr
Sfxn4
|
|
Weight adjusted by age
|
20.87 |
1 |
0 |
0 |
0 |
1.00e+00 |
Ly75
|
|
Locomotion in novel chamber
|
34.01 |
1 |
1 |
2 |
0 |
1.00e+00 |
Ly75
|
|
Food seeking constrained by brief footshock
|
15.81 |
15 |
0 |
0 |
-1 |
2.68e-152 |
Adcy3
Asxl2
Dnajc27
Dnmt3a
Drc1
Dtnb
Efr3b
Garem2
Hadha
Hadhb
Kif3c
LOC120103492
Otof
Rab10
RGD1565766
|
|
Run reversals in cocaine runway, females
|
21.72 |
1 |
0 |
0 |
0 |
1.00e+00 |
Ly75
|
|
Cu content in liver
|
31.36 |
2 |
1 |
2 |
0 |
1.00e+00 |
Cd99
LOC103694460
|
|
Fe content in liver
|
26.96 |
1 |
1 |
2 |
0 |
1.00e+00 |
LOC103694460
|
|
K content in liver
|
34.63 |
2 |
1 |
2 |
0 |
1.00e+00 |
Cd99
LOC103694460
|
|
Mn content in liver
|
32.26 |
1 |
1 |
2 |
0 |
1.00e+00 |
Ly75
|
|
Rb content in liver
|
55.75 |
2 |
2 |
4.1 |
0 |
1.00e+00 |
Cd99
LOC103694460
|
|
Sr content in liver
|
22.24 |
1 |
0 |
0 |
0 |
1.00e+00 |
LOC103694460
|