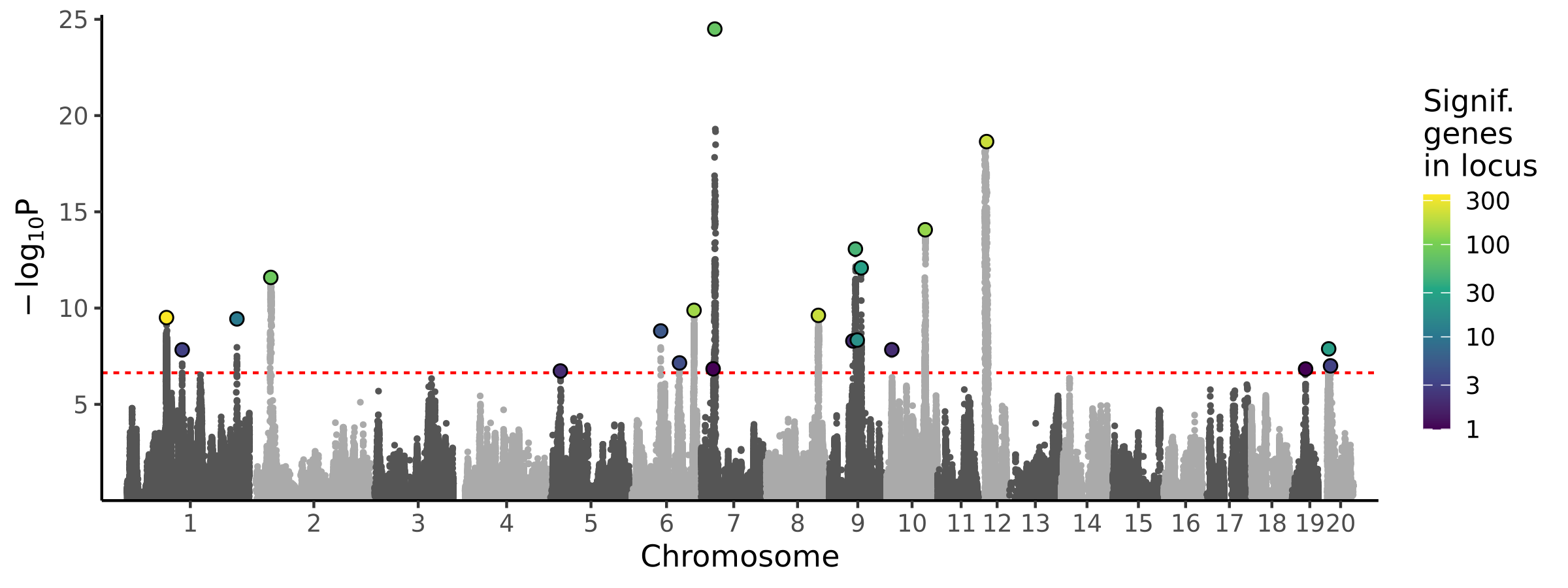
|
BMI with tail
|
14.4 |
41 |
35 |
41.7 |
-0.97 |
6.16e-113 |
Atp5mc1
B4galnt2
Calcoco2
Cavin2
Cbx1
Cdk5rap3
Copz2
Gip
Gngt2
Hoxb2
Hoxb3
Hoxb4
Hoxb9
Klhdc8b
Kpnb1
LOC102547047
LOC102547278
LOC102547955
LOC102550580
LOC108352124
LOC120095179
LOC120095180
Lrrc46
Map2
Mrpl10
Mrpl45
Nfe2l1
Npepps
Osbpl7
Phb1
Phospho1
Pnpo
Prr15l
Scrn2
Skap1
Snf8
Sp2
Tbkbp1
Tmeff2
Ube2z
Zfp652
|
|
BMI without tail
|
15.09 |
3 |
1 |
1.2 |
0 |
1.00e+00 |
Klhdc8b
LOC102553539
Mir331
|
|
Body weight
|
10.13 |
150 |
131 |
156 |
0.99 |
0.00e+00 |
Abcf1
Als2cl
Amdhd1
Apba2
Arhgef18
Arih2
Atrip
B3glct
Brca2
Camp
Camsap3
Ccdc12
Ccdc38
Ccdc51
Cd209a
Cdc25a
Cdk17
Celsr3
Cep83
Cep83os
Cers4
Cradd
Cspg5
Dclk3
Dhx16
Elavl1
Elk3
Elp6
Epm2aip1
Fam240a
Fgd6
Fkbp5
Fry
Gnl1
Hal
Hmgb1
Hsph1
Insr
Katnal1
Kif9
Kl
Klhdc8b
Klhl18
Lnc001
LOC102546827
LOC102548260
LOC102548389
LOC102548848
LOC102548950
LOC102549089
LOC102549494
LOC102552452
LOC102552504
LOC102553253
LOC102553539
LOC102554603
LOC102556485
LOC103690858
LOC108351432
LOC108351597
LOC108351781
LOC108352411
LOC120093125
LOC120093556
LOC120093559
LOC120093563
LOC120094263
LOC120095826
LOC120095830
LOC120095870
LOC120095871
LOC120095873
LOC498675
Lrrc2
Lrrc8e
Lrrfip2
Lta4h
Map2k7
Map4
Mcemp1
Mcoln1
Medag
Metap2
Mir331
Mlh1
Mrpl42
Myl3
N4bp2l1
N4bp2l2
Nbeal2
Nckipsd
Ndufa12
Nedd1
Ngp
Nme6
Nr2c1
Nradd
Ntn4
Nudt4
Pcgf2
Pcp2
Pds5b
Pet100
Pex11g
Pfkfb4
Pip4k2b
Plxnb1
Plxnc1
Pnpla6
Prr3
Prss50
Pth1r
Ptpn23
Retn
RT1-N2
RT1-N3
RT1-S3
RT1-T24-2
RT1-T24-4
Rtp3
Rxfp2
Sars2
Scap
Setd2
Shisa5
Slc26a6
Slc26a8
Slc7a1
Smarcc1
Snapc2
Snrpf
Socs2
Srcin1
Srpk1
Stard13
Stxbp2
Susd6
Timm44
Tma7
Tmcc3
Tmie
Trank1
Trappc5
Trex1
Ube2n
Uqcrc1
Vezt
Wdr95
Xab2
Zfp958
|
|
Epididymis fat weight
|
9.79 |
1 |
0 |
0 |
0 |
1.00e+00 |
Map2
|
|
Heart weight
|
17.24 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Left kidney weight
|
10.37 |
47 |
28 |
33.3 |
0.99 |
0.00e+00 |
Arhgef18
B3glct
Brca2
Camsap3
Cd209a
Cers4
Elavl1
Fry
Hmgb1
Hsph1
Insr
Katnal1
Kl
Klhdc8b
Lnc001
LOC102546827
LOC102548389
LOC102549089
LOC102549494
LOC102552452
LOC102553253
LOC103690858
LOC108352411
LOC120095826
LOC120095830
LOC120095870
LOC120095871
LOC120095873
Map2k7
Mcemp1
Mcoln1
Medag
N4bp2l1
N4bp2l2
Pcp2
Pet100
Pex11g
Pnpla6
Rxfp2
Snapc2
Stard13
Stxbp2
Timm44
Trappc5
Wdr95
Xab2
Zfp958
|
|
Right kidney weight
|
8.81 |
63 |
25 |
29.8 |
0.99 |
1.31e-291 |
Aplp1
Arhgef18
Atp4a
B3glct
Brca2
Camsap3
Cd209a
Cers4
Dmkn
Elavl1
Fry
Haus5
Hmgb1
Hspb6
Hsph1
Insr
Katnal1
Kirrel2
Kl
Klhdc8b
Lgi4
Lin37
Lnc001
LOC102546827
LOC102548389
LOC102549089
LOC102549494
LOC102552452
LOC102553253
LOC102554603
LOC103690858
LOC108351597
LOC108352411
LOC120095826
LOC120095830
LOC120095870
LOC120095871
LOC120095873
Mag
Map2k7
Mcemp1
Mcoln1
Medag
N4bp2l1
N4bp2l2
Pcp2
Pet100
Pex11g
Proser3
Psenen
Rxfp2
Slc7a1
Snapc2
Stard13
Stxbp2
Thap8
Tmem147
Trappc5
U2af1l4
Wdr95
Xab2
Zfp74
Zfp958
|
|
Tail length
|
9.24 |
247 |
119 |
141.7 |
0.98 |
0.00e+00 |
Abcf1
Acadl
Adamts6
Als2cl
Amdhd1
Ankrd44
Aox1
Aox2
Aox4
Arhgef18
Arih2
Atp5mc1
Atrip
B4galnt2
Brca2
Calcoco2
Camp
Camsap3
Capns1
Casp8
Catsperg
Cavin2
Cbx1
Ccdc12
Ccdc150
Ccdc177
Ccdc38
Ccdc51
Cd209a
Cdc25a
Cdca4
Cdk17
Cdk5rap3
Celsr3
Cenpk
Cep170b
Cep83
Cers4
Cflar
Copz2
Coq10b
Cox16
Cox7a1
Cradd
Cspg5
Cwc25
Cwc27
Dclk3
Dhx16
Dnah7
Dpf1
Elavl1
Elk3
Elp6
Epm2aip1
Erbin
Fam240a
Fam98c
Fgd6
Fry
Gip
Gngt2
Gnl1
Gtf3c3
Hecw2
Hoxb2
Hoxb3
Hoxb4
Hoxb9
Hspd1
Insr
Jag2
Kansl1l
Katnal1
Kctd18
Kif9
Kl
Klhdc8b
Klhl18
Kpnb1
Lancl1
Lnc001
LOC100912787
LOC102546726
LOC102547047
LOC102547278
LOC102547955
LOC102548389
LOC102548950
LOC102549089
LOC102550420
LOC102550580
LOC102552452
LOC102552504
LOC102553539
LOC102554603
LOC102556485
LOC103690858
LOC108351432
LOC108351597
LOC108351781
LOC108352124
LOC120093125
LOC120093556
LOC120093559
LOC120093563
LOC120094263
LOC120094685
LOC120095179
LOC120095180
LOC120095826
LOC120095871
LOC120095873
LOC120100693
LOC120100694
LOC120101075
LOC120101076
LOC120101077
LOC120101078
LOC498675
LOC686030
Lrrc2
Lrrc46
Lrrfip2
Lta4h
Maip1
Map2
Map2k7
Map4
Map4k1
Mars2
Mast4
Mcoln1
Metap2
Mir331
Mlh1
Mob4
Mrpl10
Mrpl42
Mrpl45
Mtus2
Myl3
N4bp2l1
N4bp2l2
Nbeal2
Nckipsd
Ndufa12
Nedd1
Nfe2l1
Ngp
Nln
Nme6
Npepps
Nr2c1
Nradd
Ntn4
Nudt4
Osbpl7
Pcgf2
Pcp2
Pet100
Pex11g
Pfkfb4
Pgap1
Phb1
Phospho1
Pip4k2b
Plcl1
Plxnb1
Plxnc1
Pnpla6
Pnpo
Pomp
Ppwd1
Prr15l
Prr3
Prss50
Psmd8
Pth1r
Ptpn23
Rftn2
Rgs7bp
Rpe
Rpp30
RT1-N2
RT1-N3
RT1-S3
RT1-T24-2
RT1-T24-4
Rtp3
Sars2
Scap
Scrn2
Setd2
Sf3b1
Sgtb
Shisa5
Shisal2b
Shld3
Sipa1l3
Skap1
Slc26a6
Slc39a10
Slc46a3
Slc7a1
Smarcc1
Snapc2
Snf8
Snrpf
Snx11
Socs2
Sp2
Srcin1
Srek1
Srek1ip1
Srpk1
Stard13
Stk17b
Stxbp2
Susd6
Tbkbp1
Tedc1
Timm44
Tma7
Tmcc3
Tmeff2
Tmem237
Tmie
Trank1
Trappc13
Trappc5
Trex1
Trim23
Ube2n
Ube2z
Unc80
Uqcrc1
Vezt
Xab2
Zfp14
Zfp420
Zfp566
Zfp568
Zfp652
Zfp82
Zfp84
Zfp958
|
|
Length without tail
|
11.72 |
60 |
27 |
32.1 |
0.98 |
2.12e-272 |
Apba2
Arhgef18
Brca2
Camsap3
Cd209a
Cenpk
Cers4
Dclk3
Dmkn
Elavl1
Fgd6
Fry
Haus5
Hmgb1
Hsph1
Insr
Katnal1
Kl
LOC102546827
LOC102548389
LOC102549089
LOC102549494
LOC102552452
LOC102553253
LOC102553539
LOC103690858
LOC108352411
LOC120093563
LOC120095826
LOC120095830
LOC120095871
LOC120095873
Lrrc8e
Lta4h
Mag
Map2k7
Mcemp1
Mcoln1
Medag
Mir331
Mrpl42
N4bp2l1
N4bp2l2
Ndufa12
Nr2c1
Pcp2
Pet100
Pex11g
Plxnc1
Rgs7bp
Rxfp2
Snapc2
Stard13
Stxbp2
Timm44
Trappc5
Vezt
Wdr95
Xab2
Zfp958
|
|
Liver weight, left
|
13.45 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Liver weight, right
|
13.69 |
1 |
0 |
0 |
0 |
1.00e+00 |
Srpk1
|
|
Retroperitoneal fat weight
|
6.24 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Extensor digitorum longus weight
|
8.28 |
67 |
38 |
45.2 |
0.95 |
0.00e+00 |
Ahnak2
Arhgef18
Brca2
Brf1
Btbd6
C6h14orf180
Camsap3
Cd209a
Cdca4
Cep170b
Cers4
Clba1
Crip1
Crip2
Cwc25
Cwc27
Elavl1
Fry
Gpr132
Inf2
Insr
Jag2
Kl
LOC102548389
LOC102549089
LOC102549494
LOC102552388
LOC102552452
LOC103690858
LOC108351305
LOC120095826
LOC120095871
LOC120095873
LOC120103631
LOC120103632
Lrrc8e
Map2k7
Mcemp1
Mcoln1
Mta1
N4bp2l1
N4bp2l2
Npepps
Nudt14
Pacs2
Pcgf2
Pcp2
Pet100
Pex11g
Pip4k2b
Pld4
Rgs7bp
Rxfp2
Siva1
Snapc2
Srcin1
Srek1ip1
Stard13
Stxbp2
Tedc1
Tex22
Timm44
Tmem121
Tmem179
Trappc5
Xab2
Zfp958
|
|
Soleus weight
|
7.87 |
2 |
0 |
0 |
0 |
1.00e+00 |
Car8
Klhdc8b
|
|
Tibialis anterior weight
|
7.45 |
37 |
22 |
26.2 |
0.99 |
1.23e-297 |
Arhgef18
Brca2
Camsap3
Cd209a
Cers4
Dnajb9
Elavl1
Fry
Insr
Kl
LOC102548389
LOC102549089
LOC102549494
LOC102552452
LOC103690858
LOC120095826
LOC120095871
LOC120095873
Lrrc8e
Map2k7
Mcemp1
Mcoln1
N4bp2l1
N4bp2l2
Pcp2
Pet100
Pex11g
Pnpla6
Retn
Rxfp2
Snapc2
Stard13
Stxbp2
Timm44
Trappc5
Xab2
Zfp958
|
|
Tibia length
|
7.9 |
93 |
21 |
25 |
0.99 |
0.00e+00 |
Als2cl
Amdhd1
Arih2
Atrip
Camp
Ccdc12
Ccdc38
Ccdc51
Cdc25a
Celsr3
Cenpk
Cep83
Cep83os
Coq10b
Cspg5
Dclk3
Elk3
Elp6
Epm2aip1
Erbin
Fgd6
Hspd1
Insr
Kif9
Klhdc8b
Klhl18
LOC102548848
LOC102548950
LOC102552504
LOC102553539
LOC102556485
LOC108351781
LOC120093556
LOC120093559
LOC120093563
LOC120095870
LOC120100693
LOC120100694
LOC498675
Lrrc2
Lrrfip2
Lta4h
Map4
Mars2
Mcoln1
Mir331
Mlh1
Mob4
Mrpl42
Myl3
N4bp2l1
N4bp2l2
Nbeal2
Nckipsd
Ndufa12
Nedd1
Ngp
Nln
Nme6
Nr2c1
Nradd
Ntn4
Pcp2
Pet100
Pfkfb4
Pgap1
Plxnb1
Plxnc1
Ppwd1
Prss50
Pth1r
Ptpn23
Rtp3
Scap
Setd2
Sf3b1
Sgtb
Shisa5
Shld3
Smarcc1
Snapc2
Snrpf
Socs2
Srek1
Stxbp2
Tma7
Tmcc3
Trank1
Trappc13
Trex1
Trim23
Ube2n
Vezt
|
|
Number of licking bursts
|
24.53 |
1 |
1 |
1.2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Indifference point function ln k
|
36.38 |
1 |
1 |
1.2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Indifference point function log k
|
36.38 |
1 |
1 |
1.2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Patch foraging total patch changes 6 sec
|
12.97 |
1 |
0 |
0 |
0 |
1.00e+00 |
Ahnak2
|
|
Patch foraging time to switch 0 sec
|
12.86 |
19 |
0 |
0 |
-1 |
0.00e+00 |
Ahnak2
Brf1
Btbd6
C6h14orf180
Cdca4
Cep170b
Clba1
Crip1
Crip2
Gpr132
Jag2
LOC120103631
Nudt14
Pacs2
Pld4
Siva1
Tedc1
Tex22
Tmem121
|
|
Patch foraging water rate 0 sec
|
17.19 |
25 |
22 |
26.2 |
1 |
0.00e+00 |
Ahnak2
Brf1
Btbd6
C6h14orf180
Cdca4
Cep170b
Clba1
Crip1
Crip2
Gpr132
Inf2
Jag2
LOC102552388
LOC108351305
LOC120103631
LOC120103632
Mta1
Nudt14
Pacs2
Pld4
Siva1
Tedc1
Tex22
Tmem121
Tmem179
|
|
Patch foraging water rate 12 sec
|
12.45 |
1 |
0 |
0 |
0 |
1.00e+00 |
Brf1
|
|
Patch foraging water rate 6 sec
|
13.65 |
18 |
0 |
0 |
1 |
1.98e-265 |
Adamts6
Cenpk
Cwc27
Erbin
LOC120100694
LOC120101076
LOC120101078
Mast4
Nln
Ppwd1
Rgs7bp
Sgtb
Shisal2b
Shld3
Srek1
Srek1ip1
Trappc13
Trim23
|
|
Locomotor activity
|
23.45 |
4 |
2 |
2.4 |
0.99 |
3.09e-04 |
Dnajb9
Klhdc8b
LOC103692619
Pnpla8
|
|
Locomotor testing distance
|
22.86 |
4 |
2 |
2.4 |
0.98 |
7.25e-04 |
Dnajb9
Klhdc8b
LOC103692619
Pnpla8
|
|
Locomotor testing rearing
|
14 |
1 |
0 |
0 |
0 |
1.00e+00 |
Pnpla8
|
|
Reaction time number correct
|
18.64 |
1 |
1 |
1.2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Reaction time num false alarms
|
18.55 |
1 |
0 |
0 |
0 |
1.00e+00 |
Crebbp
|
|
Reaction time num false alarms AUC
|
15.63 |
1 |
0 |
0 |
0 |
1.00e+00 |
Crebbp
|
|
Reaction time trials correct on left
|
18.64 |
1 |
1 |
1.2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Reaction time trials on left
|
19.51 |
1 |
1 |
1.2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Reaction time omissions
|
23.75 |
1 |
1 |
1.2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Reaction time false alarm rate
|
20.24 |
1 |
1 |
1.2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Reaction time premature initiations
|
13.83 |
1 |
0 |
0 |
0 |
1.00e+00 |
Srek1ip1
|
|
Reaction time trials completed
|
19.51 |
1 |
1 |
1.2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Reaction time trials AUC
|
20.65 |
1 |
1 |
1.2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Conditioned locomotion
|
28.4 |
1 |
1 |
1.2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Cocaine response before conditioning
|
38.66 |
1 |
1 |
1.2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Pavlov. Cond. magazine entry number
|
30 |
1 |
1 |
1.2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Conditioned reinforcement - inactives
|
16.77 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Intermitt. access day 15 inactive lever presses
|
30.78 |
1 |
1 |
1.2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Intermittent access terminal intake (last 3 days)
|
24.03 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Short access day 10 total inactive lever presses
|
101.65 |
1 |
1 |
1.2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Locomotion distance, session 1
|
23.98 |
1 |
1 |
1.2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Locomotion distance, session 2
|
33.15 |
1 |
1 |
1.2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Locomotion velocity, session 3
|
17.14 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Locomotion distance, session 8
|
52.78 |
1 |
1 |
1.2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Degree of sensitization distance
|
24.75 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Condit. Reinf. active minus inactive responses
|
35.59 |
1 |
1 |
1.2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Condit. Reinf. active-inactive response ratio
|
25.6 |
1 |
1 |
1.2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Condit. Reinf. active responses
|
18.23 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Incentive salience index mean
|
26.23 |
1 |
1 |
1.2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Time in familiar zone, hab. session 1
|
63.49 |
1 |
1 |
1.2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Total zone transitions, hab. session 1
|
37.38 |
1 |
1 |
1.2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Time in familiar zone, hab. session 2
|
27.1 |
1 |
1 |
1.2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Total zone transitions, hab. session 2
|
69.58 |
1 |
1 |
1.2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Total locomotion distance, hab. session 2
|
19.2 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Locomotion velocity, hab. session 2
|
21.15 |
1 |
1 |
1.2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Novel to familiar place preference ratio
|
22.33 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Novelty place preference
|
20.99 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Total zone transitions, NPP test
|
106.24 |
1 |
1 |
1.2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Total locomotion distance, NPP test
|
47.64 |
1 |
1 |
1.2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Locomotion velocity, NPP test
|
51.12 |
1 |
1 |
1.2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Pavlov. Cond. change in total contacts
|
69.48 |
1 |
1 |
1.2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Bone: apparent density
|
12.56 |
11 |
0 |
0 |
0.99 |
1.87e-36 |
Fry
Hsph1
LOC102546827
LOC102548389
LOC102549494
LOC120095871
LOC120095873
Mcemp1
Rxfp2
Trappc5
Zfp958
|
|
Bone surface
|
11.9 |
11 |
0 |
0 |
0.99 |
7.24e-38 |
Fry
Hsph1
LOC102546827
LOC102548389
LOC102549494
LOC120095871
LOC120095873
Rxfp2
Snapc2
Trappc5
Zfp958
|
|
Bone volume
|
12.34 |
9 |
0 |
0 |
1 |
4.53e-32 |
Fry
Hsph1
LOC102546827
LOC102548389
LOC102549494
LOC120095871
LOC120095873
Rxfp2
Zfp958
|
|
Bone: connectivity density
|
13.24 |
13 |
1 |
1.2 |
0.99 |
7.64e-38 |
Fry
Hsph1
LOC102546827
LOC102548389
LOC102549494
LOC120095830
LOC120095871
LOC120095873
Mcemp1
Rxfp2
Snapc2
Trappc5
Zfp958
|
|
Bone: endosteal estimation
|
8.75 |
1 |
0 |
0 |
0 |
1.00e+00 |
Slc26a8
|
|
Bone: marrow area
|
8.83 |
1 |
0 |
0 |
0 |
1.00e+00 |
Slc26a8
|
|
Bone: stiffness
|
13.41 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Bone: trabecular number
|
12.91 |
4 |
0 |
0 |
1 |
9.38e-09 |
Hsph1
LOC102546827
LOC102549494
Rxfp2
|
|
Time to tail flick, vehicle, after self-admin
|
37.57 |
1 |
1 |
1.2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Total heroin consumption
|
18.89 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Delay disc. indifference point, 0s delay
|
28.7 |
1 |
1 |
1.2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Delay disc. indifference point, 4s delay
|
41.82 |
1 |
1 |
1.2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Rest time, locomotor task time 1
|
22.55 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Distance moved, locomotor task time 1
|
31.01 |
1 |
1 |
1.2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Vertical activity count, locomotor time 1
|
49.01 |
1 |
1 |
1.2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Bouts of movement, locomotor time 2
|
19.85 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Total resting periods, locomotor time 2
|
20.22 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Rest time, locomotor task time 2
|
25.34 |
1 |
1 |
1.2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Distance moved, locomotor task time 2
|
42.39 |
1 |
1 |
1.2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Vertical activity count, locomotor time 2
|
54.92 |
1 |
1 |
1.2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Weight adjusted by age
|
30.32 |
1 |
1 |
1.2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Cd content in liver
|
20.17 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Fe content in liver
|
40.39 |
1 |
1 |
1.2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Rb content in liver
|
51.88 |
1 |
1 |
1.2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Sr content in liver
|
22.41 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|