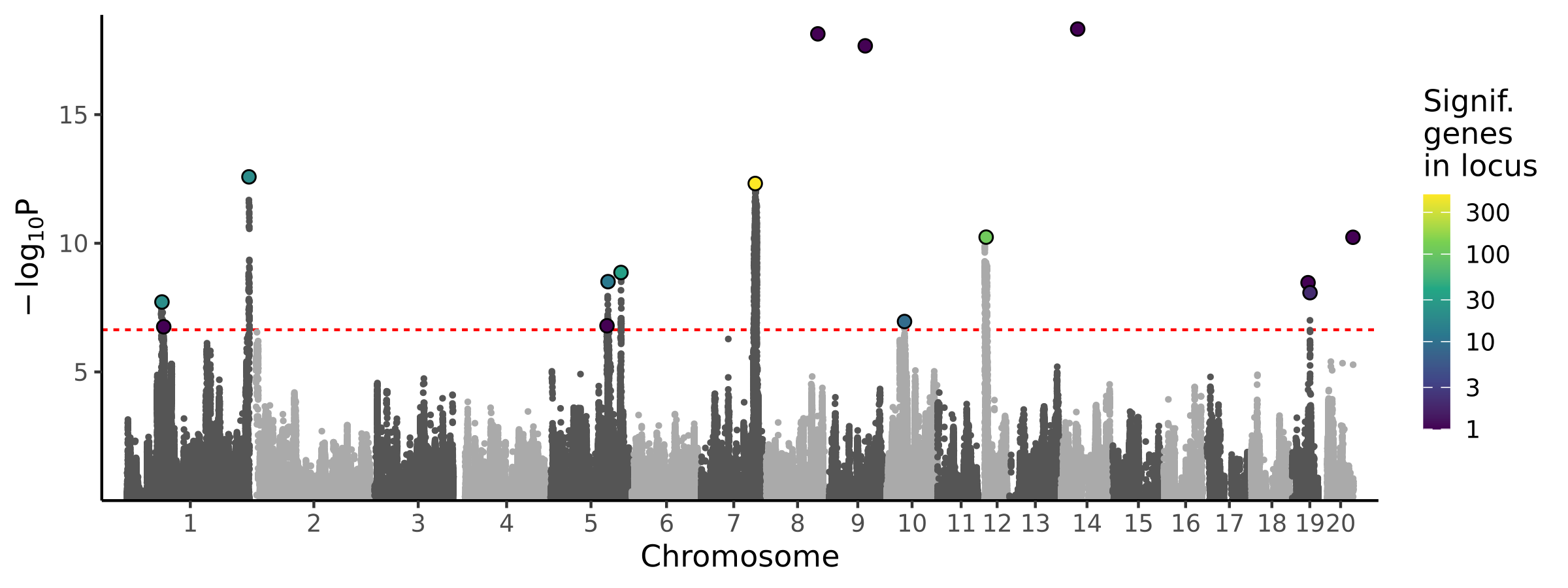
|
BMI with tail
|
65.71 |
26 |
17 |
24.6 |
0.84 |
1.33e-24 |
Cacul1
Ces2c
Dennd10
Emx2os
Eno4
Grk5
Hdac10
Lmf2
LOC120100068
LOC134485266
LOC134485267
Mapk11
Mlc1
Nanos1
Ndufb9
Ppp6r2
Prdx3
Prlhr
Rab11fip2
Retreg2
Selenoo
Sfxn4
Tafa5
Ttll8
Zfp933l1
Zfp958l1
|
|
BMI without tail
|
1509.95 |
30 |
18 |
26.1 |
-0.99 |
3.64e-82 |
Cacul1
Ces2c
Dennd10
Elavl1
Emx2os
Eno4
Fgd6
Grk5
Lcmt1
LOC102553539
LOC120100068
LOC134485266
LOC134485267
LOC134485553
Lrrc8e
Lta4h
Metap2
Nanos1
Ndufb9
Nr2c1
Pet100
Prdx3
Prlhr
Rab11fip2
Retreg2
Sfxn4
Snapc2
Vezt
Zfp933l1
Zkscan2
|
|
Body weight
|
14.15 |
117 |
111 |
160.9 |
0.99 |
0.00e+00 |
Abcf1
Alox5ap
Arhgap28
Arhgef18
Atat1
B3glct
Brca2
Cacul1
Camsap3
Cchcr1
Cd209a
Cdk17
Cers4
Ces2c
Ctxn1
Dennd10
Dhx16
Elavl1
Emx2os
Eno4
Fgd6
Flot1
Fry
Grk5
Hmgb1
Hsph1
Insr
Katnal1
Kl
Lcmt1
Lnc001
LOC102546827
LOC102549089
LOC102549494
LOC102549667
LOC102549959
LOC102551736
LOC102552452
LOC102553253
LOC102553539
LOC103690858
LOC108348331
LOC120095826
LOC120095828
LOC120095830
LOC120095867
LOC120095870
LOC120095871
LOC120095873
LOC120095877
LOC120100068
LOC120101883
LOC134481138
LOC134481141
LOC134481142
LOC134481143
LOC134481147
LOC134481148
LOC134481222
LOC134481229
LOC134481231
LOC134481239
LOC134481240
LOC134481241
LOC134481242
LOC134481243
LOC134481246
LOC134483845
LOC134483846
LOC134485266
LOC134485267
LOC134485553
Lrrc8e
Lta4h
Map2k7
Mcoln1
Medag
Metap2
N4bp2l1
N4bp2l2
Nanos1
Ndufb9
Nr2c1
Pcp2
Pds5b
Pet100
Pex11g
Polk
Prdx3
Prlhr
Pym1-ps4
Rab11fip2
Retn
RT1-CE4
RT1-O1l1
RT1-S2
RT1-T24-3
RT1-T24-4
Rxfp2
Sfxn4
Slc7a1
Snapc2
Spetex2l3
Stard13
Stxbp2
Tcf19
Tgfbr3l
Timm44
Trappc5
Uspl1
Vezt
Wdr95
Xab2
Zfp933l1
Zfp958
Zfp958l1
Zkscan2
|
|
Epididymis fat weight
|
1589.86 |
16 |
15 |
21.7 |
-0.99 |
2.05e-55 |
Cacul1
Ces2c
Dennd10
Emx2os
Grk5
LOC120100068
LOC134485266
LOC134485267
Nanos1
Ndufb9
Prdx3
Prlhr
Rab11fip2
Retreg2
Sfxn4
Zfp933l1
|
|
Glucose
|
2366.35 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Heart weight
|
147.17 |
27 |
4 |
5.8 |
0.97 |
9.60e-65 |
A4galt
Arfgap3
Cacul1
Ces2c
Cyb5r3
Cyp2d1
Cyp2d3
Cyp2d4
Cyp2d5
Dennd10
Grk5
LOC134479949
LOC134485267
Mei1
Nanos1
Nfam1
Pacsin2
Prlhr
Rab11fip2
Retreg2
Rrp7a
Serhl2
Sfxn4
Tcf20
Tspo
Ttll1
Zfp933l1
|
|
Right kidney weight
|
43.62 |
439 |
323 |
468.1 |
1 |
0.00e+00 |
A4galt
Abcf1
Acd
Aco2
Acsl6
Alox5ap
Ankdd1b
Ankra2
Ankrd54
Anxa6
Ap2s1
Apoc1
Apoc2
Apoc4
Apoe
Arfgap3
Arhgap28
Arhgap35
Arhgap8
Arhgef18
Arhgef28
Atat1
Atf4
Atxn10
Atxn1l
B3glct
Baiap2l2
Bcam
Bik
Brca2
C1h19orf81
C7h22orf23
Cabp5
Cacna1i
Cacul1
Calm3
Camsap3
Card10
Cblc
Cbx6
Cbx7
Cby1
Ccdc134
Ccdc61
Ccdc8
Ccdc9
Cchcr1
Cd209a
Cdc42ep1
Cdc42ep5
Cdc42se2
Cdk17
Cdpf1
Celsr1
Cenpm
Cerk
Cers4
Ces2c
Cgm4
Clasrp
Clptm1
Copg2
Cox6b2
Cpt1c
Cpt2
Csdc2
Csnk1e
Ctu1
Ctxn1
Cyb5r3
Cyp2d1
Cyp2d2
Cyp2d3
Cyp2d4
Cyp2d5
Czib
Dact3
Ddx17
Dennd10
Dennd6b
Desi1
Dhx16
Dmc1
Dmpk
Dmwd
Dnaaf3
Dnal4
Efcab6
Ehd2
Eif3l
Elavl1
Eml2
Emx2os
Enc1
Eno4
Enthd1
Ep300
Epn1
Eps8l1
Ercc1
Ercc2
F2rl1
Fam118a
Fbln1
Fbxo46
Fcgrt
Fgd6
Fkrp
Flot1
Fnip1
Fosb
Foxa3
Fry
Fuz
Ganc
Gcat
Gemin7
Gipr
Gm2a
Gng8
Gp6
Gpr4
Gramd4
Grap2
Grk5
Gtse1
H1f0
Hdac10
Hif3a
Hmgb1
Hs3st4
Hspbp1
Hsph1
Ift25
Insr
Iqgap2
Irgc
Ist1
Josd1
Josd2
Katnal1
Kiaa0930
Kl
Klc3
Kmt5c
L3mbtl2
Lair1
Lcmt1
Lig1
Lilrb3
Lmf2
Lnc001
LOC100362109
LOC100362400
LOC102546827
LOC102549089
LOC102549494
LOC102549667
LOC102549885
LOC102549959
LOC102550768
LOC102551027
LOC102551152
LOC102551736
LOC102552452
LOC102553253
LOC102553539
LOC102554082
LOC103690858
LOC103692950
LOC103692951
LOC103692952
LOC108348331
LOC108348449
LOC108349062
LOC108349515
LOC108349752
LOC108351520
LOC120093688
LOC120093696
LOC120093704
LOC120093705
LOC120095102
LOC120095826
LOC120095828
LOC120095830
LOC120095867
LOC120095870
LOC120095871
LOC120095873
LOC120095877
LOC120099768
LOC120099772
LOC120100068
LOC120100671
LOC120100677
LOC120101883
LOC134479757
LOC134479937
LOC134479938
LOC134479943
LOC134479944
LOC134479947
LOC134479949
LOC134481138
LOC134481141
LOC134481142
LOC134481143
LOC134481147
LOC134481148
LOC134481222
LOC134481229
LOC134481231
LOC134481239
LOC134481240
LOC134481241
LOC134481242
LOC134481243
LOC134481246
LOC134482886
LOC134482894
LOC134483641
LOC134483845
LOC134483846
LOC134485266
LOC134485267
LOC134485553
LOC134485820
LOC134485824
Lrp8
Lrrc8e
Lta4h
Maff
Map2k7
Mapk11
Mcat
Mchr1
Mcoln1
Med25
Medag
Mei1
Meis3
Mest
Metap2
Mgat3
Micall1
Micb
Mief1
Mifl2
Mill1
Mlc1
Mpped1
Mrtfa
Mzf1
N4bp2l1
N4bp2l2
Nanos1
Napa
Ncaph2
Ndc1
Ndufa6
Ndufb11b
Ndufb9
Nectin2
Nfam1
Nkpd1
Nova2
Npas1
Nptxr
Nr2c1
Nsa2
Nup160l1
Nup50
Opa3
P4ha2
Pacsin2
Parvb
Parvg
Pcp2
Pdgfb
Pds5b
Pet100
Pex11g
Phf21b
Phf5a
Pick1
Pkdrej
Pla2g6
Plxnb2
Pmfbp1
Pmm1
Pnma8a
Pnma8b
Pnma8c
Pnpla3
Podn
Podxl
Poldip3
Polk
Polr1g
Polr3h
Ppara
Ppm1n
Ppp1r13l
Ppp1r37
Ppp5c
Ppp6r1
Ppp6r2
Prdx3
Prkd2
Prlhr
Prmt1
Prr5
Ptprh
Pym1-ps4
Qpctl
Rab11fip2
Rabl2
Rbx1
Rcn3
Rdh13
Retn
Retreg2
Rpl28
Rpl3
Rrp7a
Rsph6a
RT1-CE4
RT1-O1l1
RT1-S2
RT1-T24-3
RT1-T24-4
Rtn2
Rxfp2
Samm50
Sbf1
Scaf1
Scp2
Scube1
Selenoo
Selenow
Septin3
Serhl2
Sfxn4
Sgsm3
Shank3
Shisa8
Shisal1
Six5
Slc16a8
Slc1a5
Slc25a17
Slc27a5
Slc36a1
Slc7a1
Smdt1
Smim45
Snapc2
Snrpd2
Snu13
Sox10
Spetex2l3
Srebf2
Ssbp3
St13
Stard13
Strn4
Stxbp2
Sult2a1
Sult2a6
Sult4a1
Sun2
Syce3
Sympk
Syngr1
Syt3
Syt5
Tab1
Tafa5
Tbc1d22a
Tcf19
Tcf20
Tef
Tgfbr3l
Timm44
Tmem150b
Tmem184b
Tmem86b
Tnfrsf13c
Tnip1
Tnni3
Tnnt1
Tnrc6a
Tob2
Tomm40
Trabd
Trappc5
Trappc6a
Triobp
Trmu
Tsks
Tspo
Ttc38
Ttc4
Ttll1
Ttll12
Ttll8
Ttyh1
Ube2s
Usp48
Uspl1
Utp15
Vasp
Vezt
Wbp2nl
Wdr95
Wnt7b
Xab2
Xpnpep3
Xrcc6
Zbtb40
Zbtb45
Zc3h7b
Zfp111
Zfp112
Zfp180
Zfp296
Zfp446
Zfp61
Zfp821
Zfp933l1
Zfp94
Zfp94l1
Zfp958
Zfp958l1
Zkscan2
Znf235
Znf8
Zswim9
|
|
Tail length
|
172.6 |
80 |
62 |
89.9 |
-0.94 |
7.23e-265 |
Abcf1
Arhgef18
B3glct
Brca2
Camsap3
Cdk17
Cers4
Ctxn1
Dhx16
Elavl1
Fgd6
Flot1
Fry
Hmgb1
Insr
Katnal1
Lnc001
LOC102546827
LOC102549089
LOC102549494
LOC102549959
LOC102551736
LOC102552452
LOC102553539
LOC103690858
LOC108348331
LOC108349515
LOC120095826
LOC120095828
LOC120095870
LOC120095871
LOC120095873
LOC120101883
LOC134481138
LOC134481141
LOC134481142
LOC134481143
LOC134481147
LOC134481222
LOC134481229
LOC134481231
LOC134481239
LOC134481240
LOC134481241
LOC134481242
LOC134481243
LOC134481246
LOC134483846
Lrrc8e
Lta4h
Map2k7
Mcoln1
Metap2
N4bp2l1
N4bp2l2
Nr2c1
Pcp2
Pds5b
Pet100
Pex11g
Pym1-ps4
Retn
Retreg2
RT1-CE4
RT1-O1l1
RT1-S2
RT1-T24-3
Rxfp2
Snapc2
Spetex2l3
Stard13
Stxbp2
Tgfbr3l
Timm44
Trappc5
Vezt
Wdr95
Xab2
Zfp958
Zfp958l1
|
|
Length with tail
|
24.29 |
98 |
92 |
133.3 |
-0.48 |
4.55e-40 |
Abcf1
Arhgef18
Atat1
B3glct
Brca2
Camsap3
Cchcr1
Cd209a
Cdk17
Cers4
Ctxn1
Dhx16
Elavl1
Fgd6
Flot1
Fry
Hmgb1
Hsph1
Insr
Katnal1
Kl
Lnc001
LOC102546827
LOC102549089
LOC102549494
LOC102549667
LOC102549959
LOC102551736
LOC102552452
LOC102553253
LOC102553539
LOC103690858
LOC108348331
LOC108349515
LOC120095826
LOC120095828
LOC120095830
LOC120095867
LOC120095870
LOC120095871
LOC120095873
LOC120095877
LOC120101883
LOC134481138
LOC134481141
LOC134481142
LOC134481143
LOC134481147
LOC134481148
LOC134481222
LOC134481229
LOC134481231
LOC134481239
LOC134481240
LOC134481241
LOC134481242
LOC134481243
LOC134481246
LOC134483845
LOC134483846
Lrrc8e
Lta4h
Map2k7
Mcoln1
Medag
Metap2
N4bp2l1
N4bp2l2
Ndufb9
Nr2c1
Pcp2
Pds5b
Pet100
Pex11g
Pym1-ps4
Retn
Retreg2
RT1-CE4
RT1-O1l1
RT1-S2
RT1-T24-3
RT1-T24-4
Rxfp2
Slc7a1
Snapc2
Spetex2l3
Stard13
Stxbp2
Tcf19
Tgfbr3l
Timm44
Trappc5
Uspl1
Vezt
Wdr95
Xab2
Zfp958
Zfp958l1
|
|
Length without tail
|
203.73 |
102 |
77 |
111.6 |
1 |
0.00e+00 |
Abcf1
Alox5ap
Arhgap28
Arhgef18
Atat1
B3glct
Brca2
Cacul1
Camsap3
Cchcr1
Cd209a
Cdk17
Cers4
Ctxn1
Dhx16
Elavl1
Fgd6
Flot1
Fry
Grk5
Hmgb1
Hsph1
Insr
Katnal1
Kl
Lnc001
LOC102546827
LOC102549089
LOC102549494
LOC102549667
LOC102549959
LOC102551736
LOC102552452
LOC102553253
LOC102553539
LOC103690858
LOC108348331
LOC120095826
LOC120095828
LOC120095830
LOC120095867
LOC120095870
LOC120095871
LOC120095873
LOC120095877
LOC120101883
LOC134481138
LOC134481141
LOC134481142
LOC134481143
LOC134481147
LOC134481148
LOC134481222
LOC134481229
LOC134481231
LOC134481239
LOC134481240
LOC134481241
LOC134481242
LOC134481243
LOC134481246
LOC134483846
Lrrc8e
Lta4h
Map2k7
Mcoln1
Medag
Metap2
N4bp2l1
N4bp2l2
Nanos1
Ndufb9
Nr2c1
Pcp2
Pds5b
Pet100
Pex11g
Prlhr
Pym1-ps4
Retn
Retreg2
RT1-CE4
RT1-O1l1
RT1-S2
RT1-T24-3
RT1-T24-4
Rxfp2
Sfxn4
Slc7a1
Snapc2
Spetex2l3
Stard13
Stxbp2
Tcf19
Tgfbr3l
Trappc5
Uspl1
Vezt
Wdr95
Xab2
Zfp958
Zfp958l1
|
|
Liver weight, left
|
367.37 |
12 |
6 |
8.7 |
0.99 |
1.27e-33 |
Cacul1
Ces2c
Dennd10
Emx2os
Grk5
LOC120100068
LOC134485267
Polk
Prlhr
Rab11fip2
Retreg2
Sfxn4
|
|
Liver weight, right
|
45.75 |
4 |
1 |
1.4 |
0.88 |
8.55e-03 |
Cacul1
Prlhr
Retreg2
Sfxn4
|
|
Parametrial fat weight
|
38.28 |
15 |
14 |
20.3 |
-0.45 |
1.41e-04 |
Cacul1
Ces2c
Dennd10
Emx2os
Grk5
LOC120100068
LOC134485266
LOC134485267
Nanos1
Prdx3
Prlhr
Rab11fip2
Retreg2
Sfxn4
Zfp933l1
|
|
Retroperitoneal fat weight
|
57.8 |
86 |
18 |
26.1 |
-0.86 |
1.41e-101 |
Aco2
Atf4
Cacna1i
Cacul1
Cby1
Ccdc134
Cdc42ep5
Cenpm
Ces2c
Cox6b2
Csdc2
Cyp2d1
Cyp2d5
Ddx17
Dennd10
Desi1
Dmc1
Dnal4
Emx2os
Eno4
Enthd1
Grap2
Grk5
Hspbp1
Josd1
Kmt5c
L3mbtl2
Lair1
Lig1
LOC100362109
LOC103692950
LOC108348449
LOC108351520
LOC120093696
LOC120100068
LOC134479757
LOC134479943
LOC134479947
LOC134485266
LOC134485267
Mchr1
Mei1
Mgat3
Mief1
Mzf1
Nanos1
Ndufb11b
Nfam1
Nptxr
Nup160l1
Pdgfb
Pmm1
Poldip3
Polr3h
Ppara
Prdx3
Prlhr
Ptprh
Rab11fip2
Rbx1
Rdh13
Retreg2
Rpl3
Septin3
Serhl2
Sfxn4
Shisa8
Slc25a17
Slc27a5
Smim45
Snu13
St13
Sult2a6
Sun2
Syngr1
Tab1
Tef
Tnni3
Tob2
Ttyh1
Xpnpep3
Xrcc6
Zbtb45
Zc3h7b
Zfp933l1
Znf8
|
|
Intraocular pressure
|
40071.91 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Extensor digitorum longus weight
|
18.44 |
57 |
18 |
26.1 |
-0.74 |
2.52e-67 |
Arhgef18
Brca2
Camsap3
Cd209a
Cers4
Ctxn1
Elavl1
Fry
Hs3st4
Insr
Lcmt1
Lnc001
LOC102546827
LOC102549089
LOC102549494
LOC102552452
LOC108348331
LOC120095826
LOC120095871
LOC120095873
LOC120101883
LOC134481141
LOC134481142
LOC134481143
LOC134481147
LOC134481222
LOC134481229
LOC134481231
LOC134481240
LOC134481241
LOC134481243
LOC134481246
Lrrc8e
Map2k7
Mcoln1
N4bp2l1
N4bp2l2
Pcp2
Pds5b
Pet100
Pex11g
Pym1-ps4
Retn
Retreg2
Rxfp2
Snapc2
Spetex2l3
Stard13
Stxbp2
Tgfbr3l
Timm44
Tnrc6a
Trappc5
Xab2
Zfp958
Zfp958l1
Zkscan2
|
|
Soleus weight
|
115.65 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Tibialis anterior weight
|
18.94 |
58 |
33 |
47.8 |
0.85 |
1.86e-121 |
Arhgef18
Brca2
Camsap3
Cd209a
Cers4
Ctxn1
Elavl1
Hs3st4
Insr
Lcmt1
LOC102546827
LOC102549089
LOC102549494
LOC102552452
LOC103690858
LOC108348331
LOC120095826
LOC120095871
LOC120095873
LOC120101883
LOC134481138
LOC134481141
LOC134481142
LOC134481143
LOC134481147
LOC134481229
LOC134481231
LOC134481239
LOC134481240
LOC134481242
LOC134481243
LOC134481246
Lrrc8e
Map2k7
Mcoln1
N4bp2l1
N4bp2l2
Pcp2
Pds5b
Pet100
Pex11g
Pym1-ps4
Retn
Retreg2
Rxfp2
Snapc2
Spetex2l3
Stard13
Stxbp2
Tgfbr3l
Timm44
Tnrc6a
Trappc5
Triobp
Xab2
Zfp958
Zfp958l1
Zkscan2
|
|
Tibia length
|
44.75 |
29 |
8 |
11.6 |
-0.87 |
5.32e-29 |
C1h19orf81
Camsap3
Cdk17
Ctu1
Fgd6
Insr
Josd2
LOC102553539
LOC120095867
LOC120095870
LOC134481138
LOC134481143
LOC134481229
LOC134481231
LOC134481239
LOC134481240
LOC134481242
Lta4h
Mcoln1
Metap2
Nr2c1
Pet100
Pex11g
Pym1-ps4
Retreg2
Snapc2
Spetex2l3
Stxbp2
Vezt
|
|
Number of licking bursts
|
78566.65 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Food consumed during 24 hour testing period
|
106196.53 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Times rat made contact with spout
|
305.36 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Mean time between licks in bursts
|
99757.66 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Mean num. licks in bursts
|
71307.17 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Std. dev. time between licks in bursts
|
94042.02 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Water consumed over 24 hours
|
51709.36 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Patch foraging indifference point 0 sec
|
115003.89 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Patch foraging indifference point AUC
|
44379.75 |
2 |
1 |
1.4 |
0 |
1.00e+00 |
Ndufb9
Retreg2
|
|
Indifference point function ln k
|
47350.08 |
2 |
2 |
2.9 |
0 |
1.00e+00 |
Ndufb9
Retreg2
|
|
Indifference point function log k
|
47350.08 |
2 |
2 |
2.9 |
0 |
1.00e+00 |
Ndufb9
Retreg2
|
|
Patch foraging total patch changes 0 sec
|
120264.14 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Patch foraging total patch changes 12 sec
|
5447.01 |
2 |
2 |
2.9 |
0 |
1.00e+00 |
Ndufb9
Retreg2
|
|
Patch foraging total patch changes 18 sec
|
117768.16 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Patch foraging total patch changes 24 sec
|
55715.71 |
2 |
2 |
2.9 |
0 |
1.00e+00 |
Ndufb9
Retreg2
|
|
Patch foraging total patch changes 6 sec
|
16328.34 |
2 |
2 |
2.9 |
0 |
1.00e+00 |
Ndufb9
Retreg2
|
|
Patch foraging time to switch 0 sec
|
52215.04 |
2 |
2 |
2.9 |
0 |
1.00e+00 |
Ndufb9
Retreg2
|
|
Patch foraging water rate 0 sec
|
49608.69 |
2 |
1 |
1.4 |
0 |
1.00e+00 |
Ndufb9
Retreg2
|
|
Patch foraging water rate 12 sec
|
80937.39 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Patch foraging water rate 18 sec
|
103280.07 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Patch foraging water rate 24 sec
|
4691.77 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Patch foraging water rate 6 sec
|
1993.4 |
2 |
2 |
2.9 |
0 |
1.00e+00 |
Ndufb9
Retreg2
|
|
Locomotor activity
|
12.34 |
1 |
0 |
0 |
0 |
1.00e+00 |
Retreg2
|
|
Locomotor testing distance
|
29826.01 |
2 |
1 |
1.4 |
0 |
1.00e+00 |
Ndufb9
Retreg2
|
|
Locomotor testing rearing
|
76220.77 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Light reinforcement 1
|
77630.38 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Light reinforcement 2
|
53956.03 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Reaction time number correct
|
51162.85 |
2 |
2 |
2.9 |
0 |
1.00e+00 |
Ndufb9
Retreg2
|
|
Reaction time mean minus median
|
102914.73 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Reaction time mean minus median AUC
|
69474.35 |
2 |
2 |
2.9 |
0 |
1.00e+00 |
Ndufb9
Retreg2
|
|
Reaction time num false alarms
|
57075.22 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Reaction time num false alarms AUC
|
106496.51 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Reaction time trials correct on left
|
51162.85 |
2 |
2 |
2.9 |
0 |
1.00e+00 |
Ndufb9
Retreg2
|
|
Reaction time trials on left
|
50004.69 |
2 |
2 |
2.9 |
0 |
1.00e+00 |
Ndufb9
Retreg2
|
|
Reaction time mean
|
47198.46 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Reaction time mean AUC
|
56678.81 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Median of all reaction times
|
31417.12 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Reaction time omissions
|
34267.91 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Reaction time false alarm rate
|
891.26 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Reaction time premature initiation rate
|
36877.02 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Reaction time premature initiations
|
69365.42 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Std. dev. reaction times
|
74752.64 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Reaction time trials completed
|
50004.69 |
2 |
2 |
2.9 |
0 |
1.00e+00 |
Ndufb9
Retreg2
|
|
Reaction time trials AUC
|
50572.24 |
2 |
1 |
1.4 |
0 |
1.00e+00 |
Ndufb9
Retreg2
|
|
Social response proportion
|
35302.65 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Social responses
|
5682.08 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Social time
|
44238.34 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Conditioned locomotion
|
6074.35 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Cocaine response after cond. corrected
|
32916.02 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Cocaine response after cond. not corrected
|
32133.24 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Cocaine response before conditioning
|
2800.41 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Saline control response
|
12962.78 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Condit. Reinf. active minus inactive responses
|
53410.48 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Condit. Reinf. active-inactive response ratio
|
91582.24 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Condit. Reinf. active responses
|
9691.24 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Condit. Reinf. inactive responses
|
42106.18 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Condit. Reinf. lever presses
|
15873.43 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Condit. Reinf. lever reinforcers received
|
2000.77 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Pavlov. Cond. lever latency
|
10912.53 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Pavlov. Cond. magazine entry latency
|
49418.79 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Pavlov. Cond. change in total contacts
|
114781.94 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Pavlov. Cond. index score
|
47988.08 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Pavlov. Cond. latency score
|
58476.16 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Pavlov. Cond. lever contacts
|
22069.78 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Pavlov. Cond. magazine entry number
|
16773.11 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Pavlov. Cond. intertrial magazine entries
|
104933.78 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Pavlov. Cond. lever-magazine prob. diff.
|
44871.67 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Pavlov. Cond. response bias
|
5169.79 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Conditioned reinforcement - actives
|
63842.21 |
2 |
2 |
2.9 |
0 |
1.00e+00 |
Ndufb9
Retreg2
|
|
Conditioned reinforcement - inactives
|
51690.97 |
2 |
2 |
2.9 |
0 |
1.00e+00 |
Ndufb9
Retreg2
|
|
Conditioned locomotion
|
20704.56 |
2 |
2 |
2.9 |
0 |
1.00e+00 |
Ndufb9
Retreg2
|
|
Intermittent access intake day 1-15 change
|
105309.71 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Intermittent access intake escalation
|
104778.26 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Intermittent access intake escalation 2
|
107995.31 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Intermitt. access day 1 inactive lever presses
|
78854.2 |
2 |
1 |
1.4 |
0 |
1.00e+00 |
Ndufb9
Retreg2
|
|
Intermitt. access day 15 inactive lever presses
|
69437.6 |
2 |
2 |
2.9 |
0 |
1.00e+00 |
Ndufb9
Retreg2
|
|
Intermitt. access escalation Index
|
92504.22 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Intermittent access day 1 total infusions
|
77770.79 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Intermittent access day 15 total infusions
|
65746.55 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Intermittent access terminal intake (last 3 days)
|
83205.14 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Intermittent access total infusions
|
675.9 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Intermittent access day 1 locomotion
|
52235.6 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Intermittent access day 15 total locomotion
|
88923.85 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Intermittent access total locomotion
|
711.59 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Intermittent access standard deviation
|
100960.82 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Cocaine induced anxiety
|
53447.42 |
2 |
2 |
2.9 |
0 |
1.00e+00 |
Ndufb9
Retreg2
|
|
Post-drug Anxiety
|
10242.99 |
2 |
2 |
2.9 |
0 |
1.00e+00 |
Ndufb9
Retreg2
|
|
Baseline Anxiety
|
51804.38 |
2 |
2 |
2.9 |
0 |
1.00e+00 |
Ndufb9
Retreg2
|
|
Lifetime Intake
|
6830.03 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Incentive Sensitization - Responses
|
54539.98 |
2 |
2 |
2.9 |
0 |
1.00e+00 |
Ndufb9
Retreg2
|
|
Incentive Sensitization - Breakpoint
|
47608.51 |
2 |
2 |
2.9 |
0 |
1.00e+00 |
Ndufb9
Retreg2
|
|
Progressive ratio test 1 active lever presses
|
99872.69 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Progressive ratio test 1 breakpoint
|
48129.89 |
2 |
1 |
1.4 |
0 |
1.00e+00 |
Ndufb9
Retreg2
|
|
Progressive ratio test 1 inactive lever presses
|
50376.54 |
2 |
2 |
2.9 |
0 |
1.00e+00 |
Ndufb9
Retreg2
|
|
Progressive ratio test 2 active lever presses
|
57222.6 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Progressive ratio test 2 breakpoint
|
34975.96 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Progressive ratio test 2 inactive lever presses
|
82059.21 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Total sessions with >9 infusions
|
7953.9 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Short access day 1 total inactive lever presses
|
57206.26 |
2 |
2 |
2.9 |
0 |
1.00e+00 |
Ndufb9
Retreg2
|
|
Short access day 10 total inactive lever presses
|
72387.68 |
2 |
1 |
1.4 |
0 |
1.00e+00 |
Ganc
Retreg2
|
|
Short access day 1 total infusions
|
111479.43 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Short access day 10 total infusions
|
38155.72 |
2 |
2 |
2.9 |
0 |
1.00e+00 |
Ndufb9
Retreg2
|
|
Short access total infusions
|
33946.03 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Short access day 1 locomotion
|
42043.17 |
2 |
2 |
2.9 |
0 |
1.00e+00 |
Ndufb9
Retreg2
|
|
Short access day 10 total locomotion
|
90807.81 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Short access total locomotion
|
68266.59 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Compulsive drug intake
|
65439.45 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
One hour access (0.3 mA shock)
|
9669.44 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
One hour access (shock baseline)
|
70137.44 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Number of responses in last shaping day
|
51289.19 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Context. condit. distance diff. score
|
970.59 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Locomotion velocity, session 1
|
14615.25 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Locomotion distance, session 1
|
4416.18 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Locomotion velocity, session 2
|
9358 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Locomotion distance, session 2
|
2393.66 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Locomotion velocity, session 3
|
2332.53 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Locomotion distance, session 3
|
16615.78 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Stereotopy head waving bouts, day 3
|
5866.4 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Stereotopy head waving duration, day 3
|
12131.92 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Locomotion velocity, session 7
|
1310.17 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Locomotion distance, session 7
|
3744.54 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Stereotopy head waving bouts, day 7
|
5257.18 |
2 |
2 |
2.9 |
0 |
1.00e+00 |
Ndufb9
Retreg2
|
|
Stereotopy head waving duration, day 7
|
4446.48 |
2 |
2 |
2.9 |
0 |
1.00e+00 |
Ndufb9
Retreg2
|
|
Locomotion velocity, session 8
|
1892.98 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Locomotion distance, session 8
|
673.32 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Degree of sensitization distance
|
15309.16 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Degree of sensitization stereotypy
|
8496.38 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Condit. Reinf. active minus inactive responses
|
4120.78 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Condit. Reinf. active-inactive response ratio
|
1097.66 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Condit. Reinf. active responses
|
11664.93 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Condit. Reinf. inactive responses
|
19375.3 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Incentive salience index mean
|
14781.83 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Condit. Reinf. lever presses
|
21433.81 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Time in familiar zone, hab. session 1
|
41873.03 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Time in novel zone, hab. session 1
|
35522.88 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Total zone transitions, hab. session 1
|
916.07 |
2 |
1 |
1.4 |
0 |
1.00e+00 |
Ndufb9
Retreg2
|
|
Total locomotion distance, hab. session 1
|
1687 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Locomotion velocity, hab. session 1
|
1131.06 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Time in familiar zone, hab. session 2
|
23379.25 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Time in novel zone, hab. session 2
|
24924.95 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Total zone transitions, hab. session 2
|
280.53 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Total locomotion distance, hab. session 2
|
96.8 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Locomotion velocity, hab. session 2
|
145.53 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Novel to familiar place preference ratio
|
55508.81 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Novelty place preference
|
54383.96 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Time in novel zone, NPP test
|
40844.77 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Total zone transitions, NPP test
|
4127.68 |
2 |
2 |
2.9 |
0 |
1.00e+00 |
Ndufb9
Retreg2
|
|
Total locomotion distance, NPP test
|
7114 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Locomotion velocity, NPP test
|
4041.22 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Pavlov. Cond. lever latency
|
2195.61 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Pavlov. Cond. change in total contacts
|
4539.35 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Pavlov. Cond. index score
|
1769.33 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Pavlov. Cond. latency score
|
1841.19 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Pavlov. Cond. lever contacts
|
3029.77 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Pavlov. Cond. magazine entry number
|
2162.19 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Pavlov. Cond. intertrial magazine entries
|
524.02 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Pavlov. Cond. lever-magazine prob. diff.
|
1940.27 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Pavlov. Cond. response bias
|
2606.99 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Bone: apparent density
|
1600.52 |
7 |
1 |
1.4 |
-1 |
1.95e-13 |
Fry
LOC102549494
LOC102549667
LOC120095828
LOC134481147
Retreg2
Rxfp2
|
|
Bone surface
|
6289.09 |
2 |
1 |
1.4 |
-1 |
1.25e-03 |
LOC102549494
Retreg2
|
|
Bone volume
|
8465.28 |
2 |
1 |
1.4 |
-1 |
9.78e-04 |
LOC102549494
Retreg2
|
|
Bone: connectivity density
|
429.69 |
6 |
1 |
1.4 |
-0.98 |
5.17e-12 |
LOC102549494
LOC134481147
Ndufb9
Retreg2
Rxfp2
Zfp958l1
|
|
Bone: cortical apparent density
|
45266.32 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Bone: cortical area
|
604.59 |
8 |
2 |
2.9 |
-0.98 |
1.52e-23 |
Cacul1
Ces2c
Dennd10
Grk5
Prlhr
Rab11fip2
Retreg2
Sfxn4
|
|
Bone: cortical porosity
|
1837.29 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Bone: cortical porosity
|
4807.78 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Bone: cortical thickness
|
1627.26 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Bone: cortical thickness
|
12190.6 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Bone: cortical tissue density
|
48453.42 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Bone: elastic displacement
|
29626.25 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Bone: elastic work
|
1494.59 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Bone: endosteal estimation
|
35257.61 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Bone: endosteal perimeter
|
44697.39 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Bone: final force
|
1182 |
2 |
1 |
1.4 |
-0.99 |
7.02e-05 |
Cacul1
Retreg2
|
|
Bone: final moment
|
141.81 |
4 |
1 |
1.4 |
-0.95 |
3.23e-04 |
Cacul1
Grk5
Retreg2
Sfxn4
|
|
Bone: marrow area
|
37067.95 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Bone: maximum diameter
|
12205.6 |
2 |
1 |
1.4 |
0 |
1.00e+00 |
Cacul1
Retreg2
|
|
Bone: maximum force
|
8645.79 |
2 |
1 |
1.4 |
-1 |
1.07e-03 |
Cacul1
Retreg2
|
|
Bone: maximum moment
|
6186.96 |
2 |
1 |
1.4 |
-1 |
1.46e-03 |
Cacul1
Retreg2
|
|
Bone: minimum diameter
|
17196.64 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Bone: periosteal estimation
|
2554.68 |
5 |
1 |
1.4 |
-1 |
3.85e-15 |
Cacul1
Grk5
Prlhr
Retreg2
Sfxn4
|
|
Bone: periosteal perimeter
|
1403.58 |
9 |
1 |
1.4 |
-0.99 |
9.02e-28 |
Cacul1
Ces2c
Dennd10
Grk5
LOC134485267
Rab11fip2
Retreg2
Sfxn4
Zfp933l1
|
|
Bone: post-yield work
|
50847.58 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Bone: stiffness
|
35564.91 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Bone: tissue strength
|
12264.67 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Bone: trabecular number
|
20584.56 |
2 |
2 |
2.9 |
0 |
1.00e+00 |
Ndufb9
Retreg2
|
|
Bone: trabecular spacing
|
15655.12 |
2 |
2 |
2.9 |
0 |
1.00e+00 |
Ndufb9
Retreg2
|
|
Bone: trabecular thickness
|
11097.68 |
2 |
1 |
1.4 |
0 |
1.00e+00 |
Cacul1
Retreg2
|
|
Bone: trabecular tissue density
|
26819.64 |
2 |
2 |
2.9 |
0 |
1.00e+00 |
Ndufb9
Retreg2
|
|
Breakpoint value in progressive ratio session
|
112428.06 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Distance traveled before self-admin
|
67012.59 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Distance traveled after self-admin
|
100820.36 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Delta time in closed arm before/after self-admin
|
1271.86 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Delta time in open arm before/after self-admin
|
54811.83 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Diff in mean of infusions in LGA sessions
|
54251.33 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Extinction: sum of active levers before priming
|
45801.2 |
2 |
2 |
2.9 |
0 |
1.00e+00 |
Ndufb9
Retreg2
|
|
Active lever presses in extinction session 6
|
3117.3 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Delta distance traveled before/after self-admin
|
6232.22 |
2 |
2 |
2.9 |
0 |
1.00e+00 |
Ndufb9
Retreg2
|
|
Delta ambulatory time before/after self-admin
|
48027.34 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Time in closed arm before self-admin
|
88239.28 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Time in closed arm after self-admin
|
19471.75 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Time in open arm before self-admin
|
17325.13 |
2 |
2 |
2.9 |
0 |
1.00e+00 |
Ndufb9
Retreg2
|
|
Time in open arm after self-admin
|
969.26 |
2 |
2 |
2.9 |
0 |
1.00e+00 |
Ndufb9
Retreg2
|
|
Sum of active levers in priming session hrs 5-6
|
3877.89 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Time to tail flick, vehicle, before self-admin
|
51335.42 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Time to tail flick, vehicle, after self-admin
|
39440.98 |
2 |
1 |
1.4 |
0 |
1.00e+00 |
Ndufb9
Retreg2
|
|
Time to tail flick, test, before self-admin
|
16423.04 |
2 |
2 |
2.9 |
0 |
1.00e+00 |
Ndufb9
Retreg2
|
|
Time to tail flick, test, after self-admin
|
2953.86 |
2 |
2 |
2.9 |
0 |
1.00e+00 |
Ndufb9
Retreg2
|
|
Delta time to tail flick, vehicle, before/after SA
|
36533.83 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Delta time to tail flick, test, before/after SA
|
56832.34 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Ambulatory time before self-admin
|
85547.73 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Ambulatory time after self-admin
|
88230.17 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Total heroin consumption
|
961.5 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Area under the delay curve
|
81605.23 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Delay disc. indifference point, 0s delay
|
9477.47 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Delay disc. indifference point, 16s delay
|
74803.61 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Delay disc. indifference point, 2s delay
|
825.84 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Delay disc. indifference point, 24s delay
|
99983.64 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Delay disc. indifference point, 4s delay
|
2525.7 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Delay disc. indifference point, 8s delay
|
50718.73 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Delay discount exponential model param
|
60214.95 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Delay discount hyperbolic model param
|
45715.54 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Fecal boli incidents, locomotor time 1
|
75085.82 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Change in fecal boli incidents, locomotor task
|
11754.13 |
2 |
2 |
2.9 |
0 |
1.00e+00 |
Ndufb9
Retreg2
|
|
Time >=10cm from walls, locomotor time 1
|
49255.61 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Bouts of movement, locomotor time 1
|
41590.88 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Total resting periods, locomotor time 1
|
11802.64 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Rest time, locomotor task time 1
|
37982.92 |
2 |
1 |
1.4 |
0 |
1.00e+00 |
Ndufb9
Retreg2
|
|
Distance moved, locomotor task time 1
|
53110.62 |
2 |
2 |
2.9 |
0 |
1.00e+00 |
Ndufb9
Retreg2
|
|
Vertical activity count, locomotor time 1
|
109540.42 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Time >=10cm from walls, locomotor time 2
|
80997.58 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Bouts of movement, locomotor time 2
|
87156.04 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Total resting periods, locomotor time 2
|
95047.72 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Rest time, locomotor task time 2
|
83778.18 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Distance moved, locomotor task time 2
|
34268.85 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Vertical activity count, locomotor time 2
|
138414.05 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Weight adjusted by age
|
54002.71 |
2 |
2 |
2.9 |
0 |
1.00e+00 |
Ndufb9
Retreg2
|
|
Seeking ratio, delayed vs. immediate footshock
|
49395.56 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Locomotion in novel chamber
|
8190.32 |
2 |
2 |
2.9 |
0 |
1.00e+00 |
Ndufb9
Retreg2
|
|
Locomotion in novel chamber post-restriction
|
77325.77 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Progressive ratio
|
52642.08 |
2 |
2 |
2.9 |
0 |
1.00e+00 |
Ndufb9
Retreg2
|
|
Food seeking constrained by brief footshock
|
7338.76 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Seeking ratio, punishment vs. effort
|
59148.36 |
2 |
2 |
2.9 |
0 |
1.00e+00 |
Ndufb9
Retreg2
|
|
Run reversals in cocaine runway, females
|
37174.37 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Run reversals in cocaine runway, males
|
14978.42 |
2 |
2 |
2.9 |
0 |
1.00e+00 |
Ndufb9
Retreg2
|
|
Latency to leave start box in cocaine runway
|
58575.8 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Latency to leave start box in cocaine runway, F
|
14201.52 |
2 |
2 |
2.9 |
0 |
1.00e+00 |
Ndufb9
Retreg2
|
|
Latency to leave start box in cocaine runway, M
|
7140.1 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Co content in liver
|
4721.74 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Cu content in liver
|
89.33 |
2 |
1 |
1.4 |
0 |
1.00e+00 |
Ndufb9
Retreg2
|
|
Fe content in liver
|
5656.2 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
K content in liver
|
6593.53 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Mg content in liver
|
24493.99 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Mn content in liver
|
2219.37 |
2 |
2 |
2.9 |
0 |
1.00e+00 |
Ndufb9
Retreg2
|
|
Na content in liver
|
16641.94 |
1 |
1 |
1.4 |
0 |
1.00e+00 |
Retreg2
|
|
Rb content in liver
|
241.33 |
2 |
2 |
2.9 |
0 |
1.00e+00 |
Ndufb9
Retreg2
|
|
Zn content in liver
|
7891.81 |
2 |
2 |
2.9 |
0 |
1.00e+00 |
Ndufb9
Retreg2
|