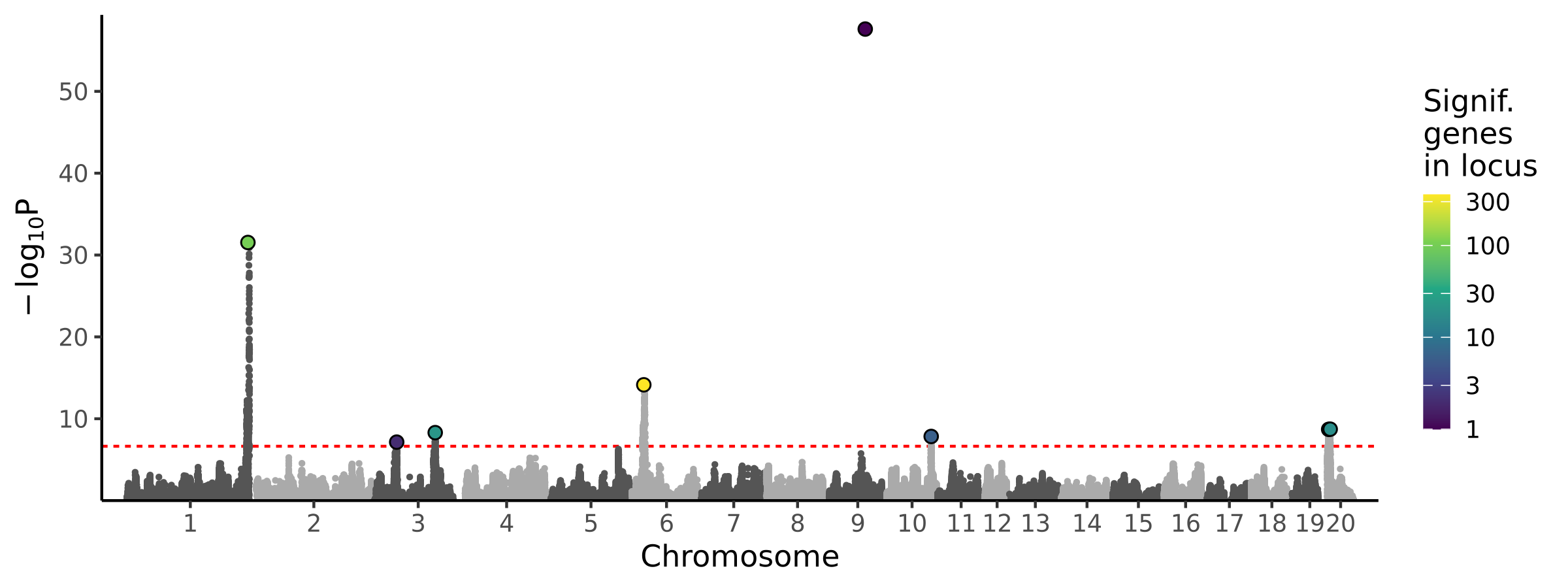
|
BMI with tail
|
16.45 |
50 |
45 |
136.4 |
0.98 |
1.00e-183 |
Ablim1
Acsl5
Afap1l2
Amz2
Arsg
Atrnl1
Axin2
Cacul1
Casp7
Ccdc186
Ces2c
Dclre1a
Dennd10
Dstn
Emx2os
Eno4
Fam204a
Fhip2a
Gpam
Grk5
Gucy2g
Habp2
Hspa12a
Kif16b
LOC102547573
LOC102550729
LOC102550782
LOC120100065
LOC120100068
LOC120101717
LOC120101719
Nanos1
Nhlrc2
Nrap
Otor
Pcsk2
Plekhs1
Prdx3
Prlhr
Rab11fip2
Rrbp1
Sfxn4
Snrpb2
Tcf7l2
Tectb
Trub1
Vti1a
Zdhhc6
Zfand2b
Zfp950
|
|
BMI without tail
|
16.67 |
24 |
15 |
45.5 |
0.99 |
1.64e-131 |
Cacul1
Ces2c
Dennd10
Dstn
Emx2os
Fam204a
Grk5
Hspa12a
Kif16b
LOC102550729
LOC120100068
LOC120101717
LOC120101719
Nanos1
Otor
Pcsk2
Prdx3
Prlhr
Rab11fip2
Rrbp1
Sfxn4
Snrpb2
Zfand2b
Zfp950
|
|
Body weight
|
16.19 |
56 |
48 |
145.5 |
0.99 |
6.54e-228 |
Ablim1
Acsl5
Adcy3
Afap1l2
Amz2
Arsg
Atrnl1
Axin2
Cacul1
Casp7
Ccdc186
Ces2c
Dclre1a
Dennd10
Dstn
Efr3b
Emx2os
Eno4
Fam204a
Fam20a
Fhip2a
Gpam
Grk5
Gucy2g
Habp2
Hspa12a
Itsn2
Kif16b
LOC102547573
LOC102550729
LOC102550782
LOC120100065
LOC120100068
LOC120101717
LOC120101719
LOC120103499
Nanos1
Nhlrc2
Nrap
Otor
Pcsk2
Plekhs1
Prdx3
Prlhr
Rab11fip2
RGD1565766
Rrbp1
Sfxn4
Snrpb2
Tcf7l2
Tectb
Trub1
Vti1a
Wipi1
Zdhhc6
Zfp950
|
|
Glucose
|
11.41 |
2 |
0 |
0 |
0 |
1.00e+00 |
LOC120098850
RT1-T24-1
|
|
Heart weight
|
18.95 |
6 |
1 |
3 |
-0.05 |
9.04e-01 |
Babam2
Cacul1
Emx2os
Rbks
Sfxn4
Zfand2b
|
|
Left kidney weight
|
11.38 |
18 |
15 |
45.5 |
0.92 |
9.54e-38 |
Acsl5
Cacul1
Ces2c
Dennd10
Emx2os
Fam204a
Gpam
Grk5
Gucy2g
LOC102550729
LOC120100068
Nanos1
Prdx3
Prlhr
Rab11fip2
Sfxn4
Zfand2b
Zfp950
|
|
Right kidney weight
|
10.46 |
29 |
15 |
45.5 |
0.91 |
5.70e-46 |
Ablim1
Acsl5
Afap1l2
Atrnl1
Cacul1
Casp7
Ccdc186
Ces2c
Dclre1a
Dennd10
Emx2os
Fam204a
Gpam
Grk5
Gucy2g
Habp2
LOC102550729
LOC120100068
Nanos1
Nhlrc2
Nrap
Plekhs1
Prdx3
Prlhr
Rab11fip2
Sfxn4
Tectb
Zfand2b
Zfp950
|
|
Tail length
|
10.76 |
18 |
3 |
9.1 |
-0.48 |
3.65e-03 |
Def6
Grm4
Ilrun
LOC103694391
LOC120098827
LOC120098967
Nudt3
Rps10
RT1-CE10
RT1-CE11
RT1-CE14
RT1-CE15
RT1-CE16
RT1-N3
RT1-T24-3
Snrpc
Spdef
Zfand2b
|
|
Length with tail
|
5.4 |
12 |
0 |
0 |
0.13 |
5.98e-01 |
Cacul1
Def6
Ilrun
Kif16b
LOC103694391
Nudt3
Otor
Sfxn4
Snrpb2
Snrpc
Spdef
Zfand2b
|
|
Length without tail
|
13.54 |
7 |
1 |
3 |
0.38 |
1.96e-01 |
Cacul1
Casp7
Emx2os
Grk5
Habp2
Prlhr
Zfand2b
|
|
Liver weight, left
|
10.94 |
3 |
0 |
0 |
1 |
5.64e-04 |
Cacul1
Grk5
Prlhr
|
|
Liver weight, right
|
12.63 |
30 |
9 |
27.3 |
0.67 |
5.83e-13 |
Cacul1
Casp7
Clip4
Def6
Dennd10
Fam204a
Gpam
Grk5
Habp2
Ilrun
LOC103694391
Nanos1
Nudt3
Pcare
Plb1
Plekhs1
Ppp1cb
Prlhr
Rab11fip2
Rbks
Rps10
Sfxn4
Snrpc
Spdef
Spdya
Togaram2
Vti1a
Zdhhc6
Zfand2b
Zfp950
|
|
Parametrial fat weight
|
14.13 |
73 |
38 |
115.2 |
0.98 |
0.00e+00 |
Abhd1
Ablim1
Acsl5
Adcy3
Afap1l2
Agbl5
Asxl2
Atrnl1
Cacul1
Casp7
Ccdc186
Cenpo
Ces2c
Dclre1a
Dennd10
Dnajc27
Dnmt3a
Drc1
Dstn
Dtnb
Efr3b
Emx2os
Eno4
Fam204a
Fam228b
Fhip2a
Fkbp1b
Garem2
Gpam
Grk5
Gucy2g
Habp2
Hadha
Hadhb
Hspa12a
Itsn2
Kcnk3
Kif16b
Kif3c
LOC102547573
LOC102550729
LOC120100065
LOC120100068
LOC120101717
LOC120101719
LOC120103492
LOC120103499
LOC120103502
Nanos1
Ncoa1
Nhlrc2
Nrap
Ost4
Otof
Otor
Pcsk2
Plekhs1
Prdx3
Prlhr
Rab10
Rab11fip2
RGD1565766
Rrbp1
Sfxn4
Slc35f6
Slc5a6
Snrpb2
Tcf7l2
Trub1
Ubxn2a
Zdhhc6
Zfand2b
Zfp950
|
|
Retroperitoneal fat weight
|
24.99 |
114 |
110 |
333.3 |
0.99 |
0.00e+00 |
Abhd1
Ablim1
Acsl5
Adcy3
Afap1l2
Agbl5
Alk
Amz2
Asxl2
Atraid
Atrnl1
Axin2
Babam2
Cacul1
Cad
Casp7
Ccdc186
Cenpo
Ces2c
Cgref1
Clip4
Dclre1a
Dennd10
Dnajc27
Dnmt3a
Dpysl5
Drc1
Dstn
Dtnb
Efr3b
Eif2b4
Emx2os
Eno4
Fam204a
Fam228b
Fhip2a
Fkbp1b
Fndc4
Fosl2
Garem2
Gckr
Gfra1
Gpam
Gpn1
Grk5
Gtf3c2
Gucy2g
Habp2
Hadha
Hadhb
Hspa12a
Ift172
Itsn2
Kcnk3
Kif16b
Kif3c
Krtcap3
Lbh
LOC102547573
LOC102548914
LOC102550729
LOC102550782
LOC120100065
LOC120100068
LOC120101717
LOC120101719
LOC120103492
LOC120103499
LOC120103502
Mpv17
Mrpl33
Nanos1
Ncoa1
Nhlrc2
Nrap
Nrbp1
Ost4
Otof
Otor
Pcare
Pcsk2
Plb1
Plekhs1
Ppm1g
Ppp1cb
Prdx3
Preb
Prlhr
Rab10
Rab11fip2
Rbks
RGD1565766
Rrbp1
Sfxn4
Slc30a3
Slc35f6
Slc4a1ap
Slc5a6
Snrpb2
Snx17
Spdya
Supt7l
Tcf7l2
Tectb
Togaram2
Trim54
Trub1
Ubxn2a
Vti1a
Zdhhc6
Zfand2b
Zfp512
Zfp513
Zfp950
|
|
Intraocular pressure
|
51.69 |
1 |
1 |
3 |
0 |
1.00e+00 |
Zfand2b
|
|
Extensor digitorum longus weight
|
4.73 |
38 |
0 |
0 |
-0.99 |
0.00e+00 |
Abhd1
Alk
Atraid
Axin2
Babam2
Cad
Cgref1
Clip4
Dpysl5
Eif2b4
Fndc4
Fosl2
Gckr
Gpn1
Gtf3c2
Ift172
Krtcap3
Lbh
LOC102548914
LOC102550782
Mpv17
Mrpl33
Nrbp1
Pcare
Plb1
Ppm1g
Ppp1cb
Preb
Rbks
Slc30a3
Slc4a1ap
Slc5a6
Snx17
Spdya
Togaram2
Trim54
Zfp512
Zfp513
|
|
Soleus weight
|
6.74 |
31 |
4 |
12.1 |
-0.36 |
7.08e-04 |
Ablim1
Acsl5
Alk
Anks1a
Atrnl1
Clip4
Def6
Emx2os
Fosl2
Gpam
Grk5
Gucy2g
Habp2
Hmga1
Ilrun
LOC103694391
LOC120098850
LOC120100068
Nudt3
Pcare
Plb1
Ppp1cb
Rbks
Rps10
Snrpc
Spdef
Spdya
Tectb
Togaram2
Trub1
Vti1a
|
|
Tibia length
|
7.21 |
5 |
0 |
0 |
0.78 |
3.58e-04 |
Fign
Kif16b
LOC120101717
Otor
Snrpb2
|
|
Number of licking bursts
|
32.5 |
1 |
1 |
3 |
0 |
1.00e+00 |
Zfand2b
|
|
Food consumed during 24 hour testing period
|
15.78 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zfand2b
|
|
Mean num. licks in bursts
|
18.13 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zfand2b
|
|
Water consumed over 24 hours
|
34.33 |
1 |
1 |
3 |
0 |
1.00e+00 |
Zfand2b
|
|
Patch foraging indifference point 0 sec
|
11.8 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zfand2b
|
|
Patch foraging indifference point AUC
|
24.2 |
1 |
1 |
3 |
0 |
1.00e+00 |
Zfand2b
|
|
Indifference point function ln k
|
54.42 |
1 |
1 |
3 |
0 |
1.00e+00 |
Zfand2b
|
|
Indifference point function log k
|
54.42 |
1 |
1 |
3 |
0 |
1.00e+00 |
Zfand2b
|
|
Patch foraging total patch changes 0 sec
|
23.48 |
1 |
1 |
3 |
0 |
1.00e+00 |
Zfand2b
|
|
Patch foraging total patch changes 18 sec
|
48.28 |
1 |
1 |
3 |
0 |
1.00e+00 |
Zfand2b
|
|
Patch foraging total patch changes 24 sec
|
77.61 |
1 |
1 |
3 |
0 |
1.00e+00 |
Zfand2b
|
|
Patch foraging total patch changes 6 sec
|
18.15 |
2 |
1 |
3 |
0 |
1.00e+00 |
Fhip2a
Zfand2b
|
|
Patch foraging time to switch 0 sec
|
33.47 |
1 |
1 |
3 |
0 |
1.00e+00 |
Zfand2b
|
|
Patch foraging water rate 0 sec
|
39.47 |
1 |
1 |
3 |
0 |
1.00e+00 |
Zfand2b
|
|
Patch foraging water rate 12 sec
|
69.49 |
1 |
1 |
3 |
0 |
1.00e+00 |
Zfand2b
|
|
Patch foraging water rate 24 sec
|
12.61 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zfand2b
|
|
Patch foraging water rate 6 sec
|
35.43 |
1 |
1 |
3 |
0 |
1.00e+00 |
Zfand2b
|
|
Locomotor activity
|
12.38 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zfand2b
|
|
Locomotor testing distance
|
47.2 |
1 |
1 |
3 |
0 |
1.00e+00 |
Zfand2b
|
|
Locomotor testing rearing
|
99.51 |
1 |
1 |
3 |
0 |
1.00e+00 |
Zfand2b
|
|
Light reinforcement 1
|
19.25 |
1 |
1 |
3 |
0 |
1.00e+00 |
Zfand2b
|
|
Reaction time number correct
|
114.79 |
1 |
1 |
3 |
0 |
1.00e+00 |
Zfand2b
|
|
Reaction time mean minus median
|
179.03 |
1 |
1 |
3 |
0 |
1.00e+00 |
Zfand2b
|
|
Reaction time mean minus median AUC
|
232.47 |
1 |
1 |
3 |
0 |
1.00e+00 |
Zfand2b
|
|
Reaction time num false alarms AUC
|
27.43 |
1 |
1 |
3 |
0 |
1.00e+00 |
Zfand2b
|
|
Reaction time trials correct on left
|
114.79 |
1 |
1 |
3 |
0 |
1.00e+00 |
Zfand2b
|
|
Reaction time trials on left
|
106.69 |
1 |
1 |
3 |
0 |
1.00e+00 |
Zfand2b
|
|
Reaction time mean
|
36.11 |
1 |
1 |
3 |
0 |
1.00e+00 |
Zfand2b
|
|
Reaction time mean AUC
|
33.75 |
1 |
1 |
3 |
0 |
1.00e+00 |
Zfand2b
|
|
Median of all reaction times
|
17.65 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zfand2b
|
|
Reaction time omissions
|
46.56 |
1 |
1 |
3 |
0 |
1.00e+00 |
Zfand2b
|
|
Reaction time premature initiations
|
41.6 |
1 |
1 |
3 |
0 |
1.00e+00 |
Zfand2b
|
|
Std. dev. reaction times
|
72.92 |
1 |
1 |
3 |
0 |
1.00e+00 |
Zfand2b
|
|
Reaction time trials completed
|
106.69 |
1 |
1 |
3 |
0 |
1.00e+00 |
Zfand2b
|
|
Reaction time trials AUC
|
108.76 |
1 |
1 |
3 |
0 |
1.00e+00 |
Zfand2b
|
|
Conditioned reinforcement - actives
|
152.54 |
1 |
1 |
3 |
0 |
1.00e+00 |
Zfand2b
|
|
Conditioned reinforcement - inactives
|
135.63 |
1 |
1 |
3 |
0 |
1.00e+00 |
Zfand2b
|
|
Intermittent access intake day 1-15 change
|
156.77 |
1 |
1 |
3 |
0 |
1.00e+00 |
Zfand2b
|
|
Intermittent access intake escalation
|
117.41 |
1 |
1 |
3 |
0 |
1.00e+00 |
Zfand2b
|
|
Intermitt. access day 1 inactive lever presses
|
169.63 |
1 |
1 |
3 |
0 |
1.00e+00 |
Zfand2b
|
|
Intermitt. access day 15 inactive lever presses
|
111.14 |
3 |
1 |
3 |
0 |
1.00e+00 |
Gpn1
Gtf3c2
Zfand2b
|
|
Intermittent access day 1 total infusions
|
71.42 |
1 |
1 |
3 |
0 |
1.00e+00 |
Zfand2b
|
|
Intermittent access day 15 total infusions
|
35.39 |
1 |
1 |
3 |
0 |
1.00e+00 |
Zfand2b
|
|
Intermittent access terminal intake (last 3 days)
|
25.82 |
1 |
1 |
3 |
0 |
1.00e+00 |
Zfand2b
|
|
Intermittent access day 1 locomotion
|
26.47 |
1 |
1 |
3 |
0 |
1.00e+00 |
Zfand2b
|
|
Intermittent access day 15 total locomotion
|
93.08 |
1 |
1 |
3 |
0 |
1.00e+00 |
Zfand2b
|
|
Intermittent access standard deviation
|
442.51 |
1 |
1 |
3 |
0 |
1.00e+00 |
Zfand2b
|
|
Cocaine induced anxiety
|
125.35 |
1 |
1 |
3 |
0 |
1.00e+00 |
Zfand2b
|
|
Baseline Anxiety
|
393.92 |
1 |
1 |
3 |
0 |
1.00e+00 |
Zfand2b
|
|
Incentive Sensitization - Responses
|
309.83 |
1 |
1 |
3 |
0 |
1.00e+00 |
Zfand2b
|
|
Incentive Sensitization - Breakpoint
|
300.45 |
1 |
1 |
3 |
0 |
1.00e+00 |
Zfand2b
|
|
Progressive ratio test 1 active lever presses
|
26.41 |
1 |
1 |
3 |
0 |
1.00e+00 |
Zfand2b
|
|
Progressive ratio test 1 breakpoint
|
19.19 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zfand2b
|
|
Progressive ratio test 2 active lever presses
|
109.22 |
1 |
1 |
3 |
0 |
1.00e+00 |
Zfand2b
|
|
Progressive ratio test 2 breakpoint
|
106.65 |
1 |
1 |
3 |
0 |
1.00e+00 |
Zfand2b
|
|
Progressive ratio test 2 inactive lever presses
|
63.47 |
1 |
1 |
3 |
0 |
1.00e+00 |
Zfand2b
|
|
Short access day 1 total inactive lever presses
|
21.93 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zfand2b
|
|
Short access day 10 total inactive lever presses
|
43.04 |
1 |
1 |
3 |
0 |
1.00e+00 |
Zfand2b
|
|
Short access day 1 locomotion
|
129.19 |
1 |
1 |
3 |
0 |
1.00e+00 |
Zfand2b
|
|
Short access day 10 total locomotion
|
432.47 |
1 |
1 |
3 |
0 |
1.00e+00 |
Zfand2b
|
|
Short access total locomotion
|
209.96 |
1 |
1 |
3 |
0 |
1.00e+00 |
Zfand2b
|
|
Compulsive drug intake
|
68.39 |
1 |
1 |
3 |
0 |
1.00e+00 |
Zfand2b
|
|
One hour access (0.3 mA shock)
|
22.58 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zfand2b
|
|
Number of responses in last shaping day
|
67.75 |
1 |
1 |
3 |
0 |
1.00e+00 |
Zfand2b
|
|
Bone volume
|
10.13 |
1 |
0 |
0 |
0 |
1.00e+00 |
Cacul1
|
|
Bone: cortical area
|
13.36 |
25 |
0 |
0 |
0.98 |
7.65e-70 |
Ablim1
Acsl5
Afap1l2
Cacul1
Casp7
Ccdc186
Ces2c
Dclre1a
Dennd10
Emx2os
Fam204a
Fhip2a
Grk5
Habp2
LOC102547573
LOC102550729
LOC120100065
LOC120100068
Nhlrc2
Plekhs1
Prlhr
Rab11fip2
Sfxn4
Trub1
Zfp950
|
|
Bone: cortical thickness
|
11.62 |
6 |
0 |
0 |
0.94 |
5.33e-05 |
Emx2os
Fam204a
LOC120100068
Prlhr
Rab11fip2
Zfp950
|
|
Bone: endosteal estimation
|
8.34 |
14 |
0 |
0 |
-1 |
3.71e-26 |
Ilrun
LOC120098827
LOC120098967
Nudt3
Rps10
RT1-CE10
RT1-CE11
RT1-CE14
RT1-CE15
RT1-CE16
RT1-N3
RT1-T24-3
Snrpc
Spdef
|
|
Bone: endosteal perimeter
|
8.34 |
1 |
0 |
0 |
0 |
1.00e+00 |
Ablim1
|
|
Bone: final force
|
11.23 |
6 |
0 |
0 |
0.99 |
1.43e-08 |
Cacul1
Grk5
Prlhr
Rab11fip2
Sfxn4
Zfp950
|
|
Bone: final moment
|
11.67 |
6 |
0 |
0 |
0.99 |
3.38e-15 |
Cacul1
Grk5
Prlhr
Rab11fip2
Sfxn4
Zfp950
|
|
Bone: marrow area
|
8.36 |
11 |
0 |
0 |
-1 |
5.95e-28 |
Ilrun
LOC120098967
Nudt3
Rps10
RT1-CE10
RT1-CE11
RT1-CE14
RT1-CE15
RT1-CE16
RT1-N3
RT1-T24-3
|
|
Bone: maximum diameter
|
10.64 |
15 |
0 |
0 |
0.99 |
8.62e-38 |
Ablim1
Acsl5
Afap1l2
Cacul1
Casp7
Ccdc186
Dclre1a
Fhip2a
Habp2
LOC102547573
LOC120100065
Nhlrc2
Nrap
Plekhs1
Trub1
|
|
Bone: maximum force
|
10.64 |
6 |
0 |
0 |
1 |
6.41e-10 |
Cacul1
Grk5
Prlhr
Rab11fip2
Sfxn4
Zfp950
|
|
Bone: maximum moment
|
11.11 |
6 |
0 |
0 |
1 |
1.30e-12 |
Cacul1
Grk5
Prlhr
Rab11fip2
Sfxn4
Zfp950
|
|
Bone: minimum diameter
|
9.1 |
1 |
0 |
0 |
0 |
1.00e+00 |
Cacul1
|
|
Bone: periosteal estimation
|
9.37 |
15 |
0 |
0 |
0.99 |
7.81e-29 |
Acsl5
Cacul1
Casp7
Dennd10
Emx2os
Fam204a
Grk5
Habp2
LOC120100065
Nhlrc2
Plekhs1
Prlhr
Rab11fip2
Sfxn4
Zfp950
|
|
Bone: periosteal perimeter
|
13.03 |
25 |
3 |
9.1 |
0.96 |
2.06e-71 |
Ablim1
Acsl5
Afap1l2
Atrnl1
Cacul1
Casp7
Ccdc186
Ces2c
Dclre1a
Dennd10
Fam204a
Fhip2a
Grk5
Habp2
LOC102547573
LOC102550729
LOC120100065
Nhlrc2
Nrap
Plekhs1
Prlhr
Rab11fip2
Sfxn4
Trub1
Zfp950
|
|
Bone: trabecular thickness
|
15.7 |
5 |
0 |
0 |
1 |
7.67e-12 |
Cacul1
Grk5
Prlhr
Sfxn4
Zfp950
|
|
Delta time in open arm before/after self-admin
|
26.67 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zfand2b
|
|
Active lever presses in extinction session 6
|
26.41 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zfand2b
|
|
Time in closed arm before self-admin
|
44.32 |
1 |
1 |
3 |
0 |
1.00e+00 |
Zfand2b
|
|
Time in open arm before self-admin
|
24.67 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zfand2b
|
|
Time to tail flick, vehicle, after self-admin
|
75.62 |
1 |
1 |
3 |
0 |
1.00e+00 |
Zfand2b
|
|
Delta time to tail flick, vehicle, before/after SA
|
74.6 |
1 |
1 |
3 |
0 |
1.00e+00 |
Zfand2b
|
|
Ambulatory time before self-admin
|
25.36 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zfand2b
|
|
Area under the delay curve
|
31.15 |
1 |
1 |
3 |
0 |
1.00e+00 |
Zfand2b
|
|
Delay disc. indifference point, 16s delay
|
69.03 |
1 |
1 |
3 |
0 |
1.00e+00 |
Zfand2b
|
|
Delay disc. indifference point, 24s delay
|
27.43 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zfand2b
|
|
Delay disc. indifference point, 8s delay
|
74.24 |
1 |
1 |
3 |
0 |
1.00e+00 |
Zfand2b
|
|
Delay discount exponential model param
|
16.14 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zfand2b
|
|
Fecal boli incidents, locomotor time 1
|
176.47 |
1 |
1 |
3 |
0 |
1.00e+00 |
Zfand2b
|
|
Time >=10cm from walls, locomotor time 1
|
79.04 |
1 |
1 |
3 |
0 |
1.00e+00 |
Zfand2b
|
|
Vertical activity count, locomotor time 1
|
97.7 |
1 |
1 |
3 |
0 |
1.00e+00 |
Zfand2b
|
|
Bouts of movement, locomotor time 2
|
23.9 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zfand2b
|
|
Total resting periods, locomotor time 2
|
36.28 |
1 |
1 |
3 |
0 |
1.00e+00 |
Zfand2b
|
|
Rest time, locomotor task time 2
|
44.91 |
1 |
1 |
3 |
0 |
1.00e+00 |
Zfand2b
|
|
Distance moved, locomotor task time 2
|
83.87 |
1 |
1 |
3 |
0 |
1.00e+00 |
Zfand2b
|
|
Food seeking constrained by brief footshock
|
15.37 |
18 |
0 |
0 |
-0.96 |
6.93e-90 |
Adcy3
Asxl2
Dnajc27
Dnmt3a
Drc1
Dtnb
Efr3b
Garem2
Hadha
Hadhb
Kif3c
LOC120103492
Ost4
Otof
Rab10
RGD1565766
Slc35f6
Zfand2b
|
|
Run reversals in cocaine runway, females
|
36.45 |
1 |
1 |
3 |
0 |
1.00e+00 |
Zfand2b
|
|
Latency to leave start box in cocaine runway
|
34.23 |
1 |
1 |
3 |
0 |
1.00e+00 |
Zfand2b
|
|
Latency to leave start box in cocaine runway, F
|
31.9 |
1 |
1 |
3 |
0 |
1.00e+00 |
Zfand2b
|
|
Se content in liver
|
15.45 |
4 |
0 |
0 |
-1 |
1.90e-03 |
Anks1a
Scube3
Snrpc
Taf11
|