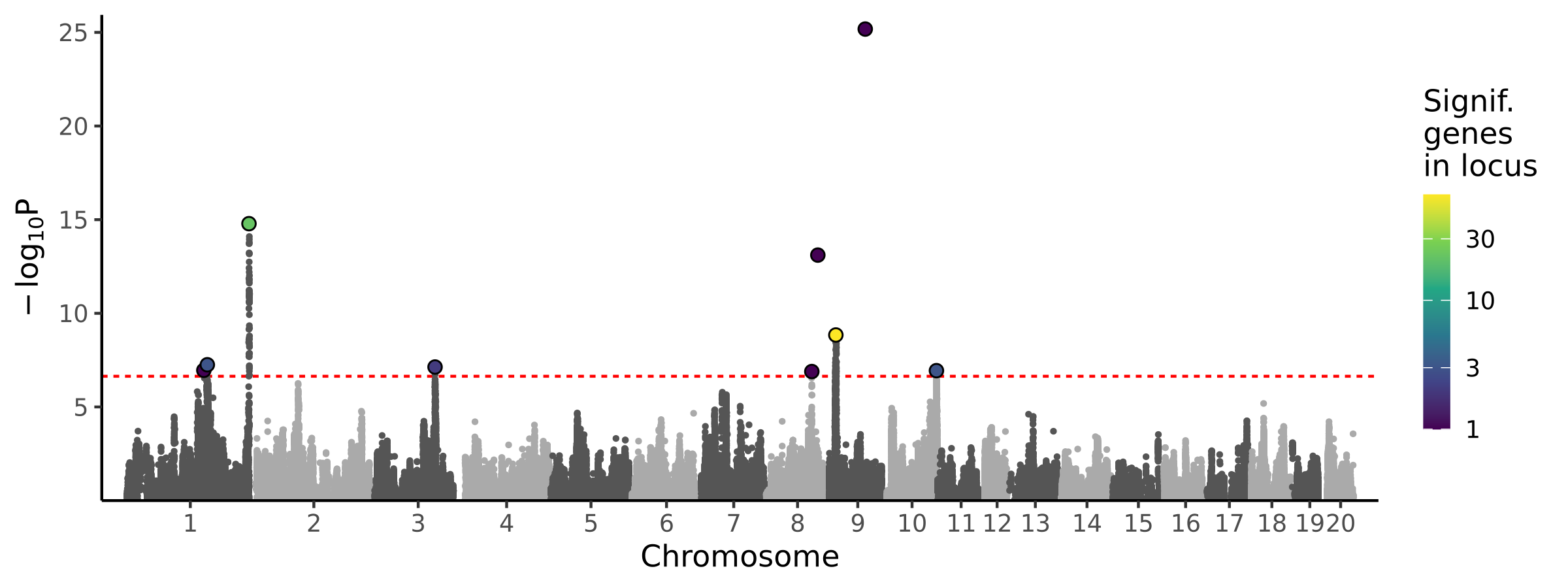
|
BMI with tail
|
15.52 |
51 |
18 |
128.6 |
0.93 |
6.63e-79 |
Aars2
Bicral
C1h11orf16
C9h6orf132
Cacul1
Capn11
Cdc5l
Ces2c
Cnpy3
Cul7
Dennd10
Emx2os
Fam204a
Gnmt
Grk5
Hsp90ab1
Kif16b
Klc4
Klhdc3
Klhdc8b
LOC100912427
LOC102550729
LOC681367
Mea1
Mrpl2
Mrps10
Nanos1
Nfkbie
Nucb2
Otor
Pex6
Ppp2r5d
Prdx3
Prlhr
Ptk7
Rab11fip2
Rrp36
Sfxn4
Slc29a1
Slc35b2
Spats1
Srf
Tcte1
Trerf1
Ubr2
Vegfa
Xrn1
Xylt1
Zfand2b
Zfp318
Zfp950
|
|
Body weight
|
17.22 |
41 |
16 |
114.3 |
0.95 |
1.29e-69 |
Bicral
C9h6orf132
Cacul1
Ces2c
Cnpy3
Cul7
Dennd10
Emx2os
Fam204a
Gnmt
Grk5
Kif16b
Klc4
Klhdc3
Klhdc8b
LOC100912427
LOC102550729
LOC681367
Mea1
Mrpl2
Mrps10
Nanos1
Nfkbie
Nucb2
Otor
Pex6
Ppp2r5d
Prdx3
Prlhr
Ptk7
Rab11fip2
Rrp36
Sfxn4
Slc29a1
Trerf1
Ubr2
Vegfa
Xrn1
Xylt1
Zfp318
Zfp950
|
|
Epididymis fat weight
|
37.04 |
16 |
15 |
107.1 |
1 |
8.27e-82 |
Cacul1
Ces2c
Dennd10
Emx2os
Fam204a
Grk5
Kif16b
LOC102550729
Nanos1
Otor
Prdx3
Prlhr
Rab11fip2
Sfxn4
Zfand2b
Zfp950
|
|
Glucose
|
9.83 |
8 |
0 |
0 |
1 |
2.38e-35 |
Cacul1
Ces2c
Dennd10
Fam204a
Grk5
LOC102550729
Sfxn4
Zfp950
|
|
Heart weight
|
12.26 |
13 |
1 |
7.1 |
0.82 |
2.00e-12 |
Cacul1
Ces2c
Dennd10
Emx2os
Fam204a
Grk5
Klhdc8b
LOC102550729
Prlhr
Rab11fip2
Sfxn4
Zfand2b
Zfp950
|
|
Left kidney weight
|
11.58 |
17 |
15 |
107.1 |
0.92 |
9.28e-33 |
Cacul1
Ces2c
Dennd10
Emx2os
Fam204a
Grk5
Klhdc8b
LOC102550729
Nanos1
Nucb2
Prdx3
Prlhr
Rab11fip2
Sfxn4
Xylt1
Zfand2b
Zfp950
|
|
Right kidney weight
|
11.38 |
17 |
15 |
107.1 |
0.9 |
9.65e-28 |
Cacul1
Ces2c
Dennd10
Emx2os
Fam204a
Grk5
Klhdc8b
LOC102550729
Nanos1
Nucb2
Prdx3
Prlhr
Rab11fip2
Sfxn4
Xylt1
Zfand2b
Zfp950
|
|
Tail length
|
46.46 |
3 |
1 |
7.1 |
0 |
1.00e+00 |
Klhdc8b
Nucb2
Zfand2b
|
|
Length with tail
|
4.18 |
10 |
1 |
7.1 |
0.99 |
5.37e-28 |
Cacul1
Ces2c
Dennd10
Grk5
Klhdc8b
Nanos1
Prlhr
Sfxn4
Zfand2b
Zfp950
|
|
Length without tail
|
11.08 |
11 |
1 |
7.1 |
0.42 |
7.63e-02 |
Aars2
Cacul1
Emx2os
Fam204a
Grk5
Nfkbie
Prlhr
Rab11fip2
Sfxn4
Zfand2b
Zfp950
|
|
Liver weight, left
|
9.96 |
9 |
0 |
0 |
0.76 |
2.81e-04 |
C1h11orf16
Cacul1
Dennd10
Fam204a
Grk5
Klhdc8b
Prlhr
Rab11fip2
Sfxn4
|
|
Liver weight, right
|
11.74 |
12 |
6 |
42.9 |
0.99 |
8.55e-58 |
Cacul1
Ces2c
Dennd10
Fam204a
Grk5
LOC102550729
Nanos1
Prlhr
Rab11fip2
Sfxn4
Zfand2b
Zfp950
|
|
Parametrial fat weight
|
20.75 |
31 |
15 |
107.1 |
0.96 |
1.22e-58 |
Aars2
Cacul1
Ces2c
Cnpy3
Cul7
Dennd10
Emx2os
Fam204a
Gnmt
Grk5
Kif16b
Klc4
Klhdc3
LOC102550729
Mea1
Mrpl2
Mrps10
Nanos1
Nucb2
Otor
Pex6
Prdx3
Prlhr
Rab11fip2
Rrp36
Sfxn4
Slc29a1
Spats1
Trerf1
Zfand2b
Zfp950
|
|
Retroperitoneal fat weight
|
41.22 |
17 |
15 |
107.1 |
0.99 |
4.35e-60 |
Cacul1
Ces2c
Dennd10
Emx2os
Fam204a
Grk5
Kif16b
Klhdc8b
LOC102550729
Nanos1
Otor
Prdx3
Prlhr
Rab11fip2
Sfxn4
Zfand2b
Zfp950
|
|
Intraocular pressure
|
51.69 |
1 |
1 |
7.1 |
0 |
1.00e+00 |
Zfand2b
|
|
Extensor digitorum longus weight
|
3.75 |
3 |
0 |
0 |
0 |
1.00e+00 |
Emx2os
Sfxn4
Xylt1
|
|
Soleus weight
|
5.45 |
6 |
0 |
0 |
1 |
2.07e-10 |
Ces2c
Grk5
Klhdc8b
Nanos1
Rab11fip2
Zfp950
|
|
Tibialis anterior weight
|
4.68 |
2 |
0 |
0 |
0 |
1.00e+00 |
Nucb2
Xylt1
|
|
Tibia length
|
10.96 |
1 |
1 |
7.1 |
0 |
1.00e+00 |
Klhdc8b
|
|
Number of licking bursts
|
28.51 |
2 |
2 |
14.3 |
0 |
1.00e+00 |
Klhdc8b
Zfand2b
|
|
Food consumed during 24 hour testing period
|
15.78 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zfand2b
|
|
Mean num. licks in bursts
|
14.92 |
2 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
Zfand2b
|
|
Std. dev. time between licks in bursts
|
12.12 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zfand2b
|
|
Water consumed over 24 hours
|
34.33 |
1 |
1 |
7.1 |
0 |
1.00e+00 |
Zfand2b
|
|
Patch foraging indifference point 0 sec
|
10.05 |
2 |
0 |
0 |
0 |
1.00e+00 |
Emx2os
Zfand2b
|
|
Patch foraging indifference point AUC
|
24.2 |
1 |
1 |
7.1 |
0 |
1.00e+00 |
Zfand2b
|
|
Indifference point function ln k
|
45.4 |
2 |
2 |
14.3 |
0 |
1.00e+00 |
Klhdc8b
Zfand2b
|
|
Indifference point function log k
|
45.4 |
2 |
2 |
14.3 |
0 |
1.00e+00 |
Klhdc8b
Zfand2b
|
|
Patch foraging total patch changes 0 sec
|
10.1 |
10 |
1 |
7.1 |
1 |
8.28e-15 |
Cacul1
Emx2os
Fam204a
Grk5
Klhdc8b
Prlhr
Rab11fip2
Sfxn4
Zfand2b
Zfp950
|
|
Patch foraging total patch changes 18 sec
|
48.28 |
1 |
1 |
7.1 |
0 |
1.00e+00 |
Zfand2b
|
|
Patch foraging total patch changes 24 sec
|
77.61 |
1 |
1 |
7.1 |
0 |
1.00e+00 |
Zfand2b
|
|
Patch foraging total patch changes 6 sec
|
13.28 |
4 |
1 |
7.1 |
0.97 |
2.94e-02 |
Emx2os
Prlhr
Rab11fip2
Zfand2b
|
|
Patch foraging time to switch 0 sec
|
15.3 |
4 |
1 |
7.1 |
1 |
1.09e-03 |
Klhdc3
LOC681367
Rrp36
Zfand2b
|
|
Patch foraging water rate 0 sec
|
23.88 |
2 |
1 |
7.1 |
0 |
1.00e+00 |
Klhdc3
Zfand2b
|
|
Patch foraging water rate 12 sec
|
69.49 |
1 |
1 |
7.1 |
0 |
1.00e+00 |
Zfand2b
|
|
Patch foraging water rate 24 sec
|
12.61 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zfand2b
|
|
Patch foraging water rate 6 sec
|
35.43 |
1 |
1 |
7.1 |
0 |
1.00e+00 |
Zfand2b
|
|
Locomotor activity
|
35.08 |
2 |
1 |
7.1 |
0 |
1.00e+00 |
Klhdc8b
Zfand2b
|
|
Locomotor testing distance
|
55.86 |
2 |
2 |
14.3 |
0 |
1.00e+00 |
Klhdc8b
Zfand2b
|
|
Locomotor testing rearing
|
99.51 |
1 |
1 |
7.1 |
0 |
1.00e+00 |
Zfand2b
|
|
Light reinforcement 1
|
19.25 |
1 |
1 |
7.1 |
0 |
1.00e+00 |
Zfand2b
|
|
Reaction time number correct
|
66.72 |
2 |
2 |
14.3 |
0 |
1.00e+00 |
Klhdc8b
Zfand2b
|
|
Reaction time mean minus median
|
98.07 |
2 |
1 |
7.1 |
0 |
1.00e+00 |
C1h11orf16
Zfand2b
|
|
Reaction time mean minus median AUC
|
122.84 |
2 |
1 |
7.1 |
0 |
1.00e+00 |
C1h11orf16
Zfand2b
|
|
Reaction time num false alarms AUC
|
27.43 |
1 |
1 |
7.1 |
0 |
1.00e+00 |
Zfand2b
|
|
Reaction time trials correct on left
|
66.72 |
2 |
2 |
14.3 |
0 |
1.00e+00 |
Klhdc8b
Zfand2b
|
|
Reaction time trials on left
|
63.1 |
2 |
2 |
14.3 |
0 |
1.00e+00 |
Klhdc8b
Zfand2b
|
|
Reaction time mean
|
36.11 |
1 |
1 |
7.1 |
0 |
1.00e+00 |
Zfand2b
|
|
Reaction time mean AUC
|
33.75 |
1 |
1 |
7.1 |
0 |
1.00e+00 |
Zfand2b
|
|
Median of all reaction times
|
14.09 |
2 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
Zfand2b
|
|
Reaction time omissions
|
35.15 |
2 |
2 |
14.3 |
0 |
1.00e+00 |
Klhdc8b
Zfand2b
|
|
Reaction time false alarm rate
|
20.24 |
1 |
1 |
7.1 |
0 |
1.00e+00 |
Klhdc8b
|
|
Reaction time premature initiations
|
41.6 |
1 |
1 |
7.1 |
0 |
1.00e+00 |
Zfand2b
|
|
Std. dev. reaction times
|
42.37 |
2 |
1 |
7.1 |
0 |
1.00e+00 |
C1h11orf16
Zfand2b
|
|
Reaction time trials completed
|
63.1 |
2 |
2 |
14.3 |
0 |
1.00e+00 |
Klhdc8b
Zfand2b
|
|
Reaction time trials AUC
|
64.7 |
2 |
2 |
14.3 |
0 |
1.00e+00 |
Klhdc8b
Zfand2b
|
|
Conditioned locomotion
|
28.4 |
1 |
1 |
7.1 |
0 |
1.00e+00 |
Klhdc8b
|
|
Cocaine response after cond. corrected
|
12.71 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zfand2b
|
|
Cocaine response after cond. not corrected
|
16.6 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Cocaine response before conditioning
|
38.66 |
1 |
1 |
7.1 |
0 |
1.00e+00 |
Klhdc8b
|
|
Pavlov. Cond. magazine entry number
|
30 |
1 |
1 |
7.1 |
0 |
1.00e+00 |
Klhdc8b
|
|
Conditioned reinforcement - actives
|
152.54 |
1 |
1 |
7.1 |
0 |
1.00e+00 |
Zfand2b
|
|
Conditioned reinforcement - inactives
|
76.2 |
2 |
1 |
7.1 |
0 |
1.00e+00 |
Klhdc8b
Zfand2b
|
|
Conditioned locomotion
|
13.93 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zfand2b
|
|
Intermittent access intake day 1-15 change
|
156.77 |
1 |
1 |
7.1 |
0 |
1.00e+00 |
Zfand2b
|
|
Intermittent access intake escalation
|
117.41 |
1 |
1 |
7.1 |
0 |
1.00e+00 |
Zfand2b
|
|
Intermittent access intake escalation 2
|
17.78 |
1 |
0 |
0 |
0 |
1.00e+00 |
C1h11orf16
|
|
Intermitt. access day 1 inactive lever presses
|
91.66 |
2 |
1 |
7.1 |
0 |
1.00e+00 |
Klhdc8b
Zfand2b
|
|
Intermitt. access day 15 inactive lever presses
|
162.52 |
2 |
2 |
14.3 |
0 |
1.00e+00 |
Klhdc8b
Zfand2b
|
|
Intermittent access day 1 total infusions
|
71.42 |
1 |
1 |
7.1 |
0 |
1.00e+00 |
Zfand2b
|
|
Intermittent access day 15 total infusions
|
24.8 |
2 |
1 |
7.1 |
0 |
1.00e+00 |
Klhdc8b
Zfand2b
|
|
Intermittent access terminal intake (last 3 days)
|
24.92 |
2 |
1 |
7.1 |
0 |
1.00e+00 |
Klhdc8b
Zfand2b
|
|
Intermittent access total infusions
|
15.45 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Intermittent access day 1 locomotion
|
26.47 |
1 |
1 |
7.1 |
0 |
1.00e+00 |
Zfand2b
|
|
Intermittent access day 15 total locomotion
|
93.08 |
1 |
1 |
7.1 |
0 |
1.00e+00 |
Zfand2b
|
|
Intermittent access total locomotion
|
12.43 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zfand2b
|
|
Intermittent access standard deviation
|
229.66 |
2 |
1 |
7.1 |
0 |
1.00e+00 |
Klhdc8b
Zfand2b
|
|
Cocaine induced anxiety
|
69.5 |
2 |
1 |
7.1 |
0 |
1.00e+00 |
Klhdc8b
Zfand2b
|
|
Baseline Anxiety
|
393.92 |
1 |
1 |
7.1 |
0 |
1.00e+00 |
Zfand2b
|
|
Incentive Sensitization - Responses
|
309.83 |
1 |
1 |
7.1 |
0 |
1.00e+00 |
Zfand2b
|
|
Incentive Sensitization - Breakpoint
|
300.45 |
1 |
1 |
7.1 |
0 |
1.00e+00 |
Zfand2b
|
|
Progressive ratio test 1 active lever presses
|
26.41 |
1 |
1 |
7.1 |
0 |
1.00e+00 |
Zfand2b
|
|
Progressive ratio test 1 breakpoint
|
19.19 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zfand2b
|
|
Progressive ratio test 1 inactive lever presses
|
16.55 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Progressive ratio test 2 active lever presses
|
109.22 |
1 |
1 |
7.1 |
0 |
1.00e+00 |
Zfand2b
|
|
Progressive ratio test 2 breakpoint
|
106.65 |
1 |
1 |
7.1 |
0 |
1.00e+00 |
Zfand2b
|
|
Progressive ratio test 2 inactive lever presses
|
63.47 |
1 |
1 |
7.1 |
0 |
1.00e+00 |
Zfand2b
|
|
Total sessions with >9 infusions
|
15.19 |
2 |
0 |
0 |
0 |
1.00e+00 |
C1h11orf16
Klhdc8b
|
|
Short access day 1 total inactive lever presses
|
17.49 |
2 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
Zfand2b
|
|
Short access day 10 total inactive lever presses
|
72.35 |
2 |
2 |
14.3 |
0 |
1.00e+00 |
Klhdc8b
Zfand2b
|
|
Short access day 10 total infusions
|
13.17 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Short access day 1 locomotion
|
129.19 |
1 |
1 |
7.1 |
0 |
1.00e+00 |
Zfand2b
|
|
Short access day 10 total locomotion
|
432.47 |
1 |
1 |
7.1 |
0 |
1.00e+00 |
Zfand2b
|
|
Short access total locomotion
|
209.96 |
1 |
1 |
7.1 |
0 |
1.00e+00 |
Zfand2b
|
|
Compulsive drug intake
|
68.39 |
1 |
1 |
7.1 |
0 |
1.00e+00 |
Zfand2b
|
|
One hour access (0.3 mA shock)
|
18.01 |
2 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
Zfand2b
|
|
Number of responses in last shaping day
|
67.75 |
1 |
1 |
7.1 |
0 |
1.00e+00 |
Zfand2b
|
|
Locomotion distance, session 1
|
23.98 |
1 |
1 |
7.1 |
0 |
1.00e+00 |
Klhdc8b
|
|
Locomotion velocity, session 2
|
14.35 |
1 |
0 |
0 |
0 |
1.00e+00 |
Trerf1
|
|
Locomotion distance, session 2
|
33.15 |
1 |
1 |
7.1 |
0 |
1.00e+00 |
Klhdc8b
|
|
Locomotion velocity, session 3
|
17.14 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Locomotion distance, session 3
|
14.51 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Locomotion distance, session 8
|
52.78 |
1 |
1 |
7.1 |
0 |
1.00e+00 |
Klhdc8b
|
|
Degree of sensitization distance
|
24.75 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Condit. Reinf. active minus inactive responses
|
35.59 |
1 |
1 |
7.1 |
0 |
1.00e+00 |
Klhdc8b
|
|
Condit. Reinf. active-inactive response ratio
|
25.6 |
1 |
1 |
7.1 |
0 |
1.00e+00 |
Klhdc8b
|
|
Condit. Reinf. active responses
|
18.23 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Incentive salience index mean
|
26.23 |
1 |
1 |
7.1 |
0 |
1.00e+00 |
Klhdc8b
|
|
Condit. Reinf. lever presses
|
10.81 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Time in familiar zone, hab. session 1
|
63.49 |
1 |
1 |
7.1 |
0 |
1.00e+00 |
Klhdc8b
|
|
Total zone transitions, hab. session 1
|
37.38 |
1 |
1 |
7.1 |
0 |
1.00e+00 |
Klhdc8b
|
|
Total locomotion distance, hab. session 1
|
10.76 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Locomotion velocity, hab. session 1
|
10.22 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Time in familiar zone, hab. session 2
|
27.1 |
1 |
1 |
7.1 |
0 |
1.00e+00 |
Klhdc8b
|
|
Total zone transitions, hab. session 2
|
69.58 |
1 |
1 |
7.1 |
0 |
1.00e+00 |
Klhdc8b
|
|
Total locomotion distance, hab. session 2
|
19.2 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Locomotion velocity, hab. session 2
|
21.15 |
1 |
1 |
7.1 |
0 |
1.00e+00 |
Klhdc8b
|
|
Novel to familiar place preference ratio
|
22.33 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Novelty place preference
|
20.99 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Time in novel zone, NPP test
|
17.06 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Total zone transitions, NPP test
|
41.93 |
3 |
1 |
7.1 |
0 |
1.00e+00 |
Klhdc3
Klhdc8b
Rrp36
|
|
Total locomotion distance, NPP test
|
28.8 |
2 |
1 |
7.1 |
0 |
1.00e+00 |
Klhdc8b
Xrn1
|
|
Locomotion velocity, NPP test
|
51.12 |
1 |
1 |
7.1 |
0 |
1.00e+00 |
Klhdc8b
|
|
Pavlov. Cond. change in total contacts
|
69.48 |
1 |
1 |
7.1 |
0 |
1.00e+00 |
Klhdc8b
|
|
Pavlov. Cond. index score
|
14.07 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Pavlov. Cond. magazine entry number
|
12.23 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Pavlov. Cond. lever-magazine prob. diff.
|
14.82 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Bone: apparent density
|
8.24 |
1 |
0 |
0 |
0 |
1.00e+00 |
Cacul1
|
|
Bone volume
|
9.37 |
5 |
0 |
0 |
1 |
1.18e-12 |
Cacul1
Grk5
Prlhr
Rab11fip2
Sfxn4
|
|
Bone: cortical apparent density
|
11.41 |
4 |
0 |
0 |
-1 |
1.27e-05 |
C1h11orf16
Cacul1
Dennd10
Sfxn4
|
|
Bone: cortical area
|
13.06 |
12 |
0 |
0 |
1 |
3.63e-83 |
Cacul1
Ces2c
Dennd10
Emx2os
Fam204a
Grk5
LOC102550729
Nanos1
Prlhr
Rab11fip2
Sfxn4
Zfp950
|
|
Bone: cortical thickness
|
9.98 |
9 |
0 |
0 |
0.99 |
3.07e-17 |
Cacul1
Ces2c
Emx2os
Fam204a
Grk5
Prlhr
Rab11fip2
Sfxn4
Zfp950
|
|
Bone: cortical thickness
|
12.52 |
1 |
0 |
0 |
0 |
1.00e+00 |
Grk5
|
|
Bone: cortical tissue density
|
11.17 |
6 |
0 |
0 |
-0.99 |
5.25e-23 |
C1h11orf16
Cacul1
Ces2c
Dennd10
Grk5
Sfxn4
|
|
Bone: final force
|
9.51 |
12 |
0 |
0 |
1 |
2.29e-53 |
Cacul1
Ces2c
Dennd10
Emx2os
Fam204a
Grk5
LOC102550729
Nanos1
Prlhr
Rab11fip2
Sfxn4
Zfp950
|
|
Bone: final moment
|
9.93 |
12 |
0 |
0 |
1 |
7.37e-60 |
Cacul1
Ces2c
Dennd10
Emx2os
Fam204a
Grk5
LOC102550729
Nanos1
Prlhr
Rab11fip2
Sfxn4
Zfp950
|
|
Bone: maximum diameter
|
8.17 |
5 |
0 |
0 |
1 |
7.21e-19 |
Cacul1
Dennd10
Grk5
Prlhr
Sfxn4
|
|
Bone: maximum force
|
9.46 |
9 |
0 |
0 |
0.99 |
1.98e-24 |
Cacul1
Emx2os
Fam204a
Grk5
Nanos1
Prlhr
Rab11fip2
Sfxn4
Zfp950
|
|
Bone: maximum moment
|
9.73 |
10 |
0 |
0 |
1 |
1.86e-26 |
Cacul1
Dennd10
Emx2os
Fam204a
Grk5
Nanos1
Prlhr
Rab11fip2
Sfxn4
Zfp950
|
|
Bone: minimum diameter
|
7.55 |
9 |
0 |
0 |
1 |
1.51e-43 |
Cacul1
Ces2c
Dennd10
Emx2os
Grk5
LOC102550729
Prlhr
Sfxn4
Zfp950
|
|
Bone: periosteal estimation
|
8.51 |
12 |
0 |
0 |
1 |
1.04e-77 |
Cacul1
Ces2c
Dennd10
Emx2os
Fam204a
Grk5
LOC102550729
Nanos1
Prlhr
Rab11fip2
Sfxn4
Zfp950
|
|
Bone: periosteal perimeter
|
11.13 |
12 |
0 |
0 |
1 |
6.20e-64 |
Cacul1
Ces2c
Dennd10
Emx2os
Fam204a
Grk5
LOC102550729
Nanos1
Prlhr
Rab11fip2
Sfxn4
Zfp950
|
|
Bone: stiffness
|
13.41 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Bone: tissue strength
|
14.55 |
1 |
0 |
0 |
0 |
1.00e+00 |
C1h11orf16
|
|
Bone: trabecular number
|
11.4 |
1 |
0 |
0 |
0 |
1.00e+00 |
C1h11orf16
|
|
Bone: trabecular spacing
|
15.64 |
1 |
0 |
0 |
0 |
1.00e+00 |
C1h11orf16
|
|
Bone: trabecular thickness
|
13.35 |
11 |
0 |
0 |
1 |
1.67e-66 |
Cacul1
Ces2c
Dennd10
Emx2os
Fam204a
Grk5
LOC102550729
Prlhr
Rab11fip2
Sfxn4
Zfp950
|
|
Breakpoint value in progressive ratio session
|
15.73 |
1 |
0 |
0 |
0 |
1.00e+00 |
Nucb2
|
|
Distance traveled before self-admin
|
17.44 |
1 |
0 |
0 |
0 |
1.00e+00 |
C1h11orf16
|
|
Delta time in open arm before/after self-admin
|
26.67 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zfand2b
|
|
Active lever presses in extinction session 6
|
26.41 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zfand2b
|
|
Time in closed arm before self-admin
|
44.32 |
1 |
1 |
7.1 |
0 |
1.00e+00 |
Zfand2b
|
|
Time in open arm before self-admin
|
24.67 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zfand2b
|
|
Time to tail flick, vehicle, after self-admin
|
56.6 |
2 |
2 |
14.3 |
0 |
1.00e+00 |
Klhdc8b
Zfand2b
|
|
Delta time to tail flick, vehicle, before/after SA
|
44.54 |
2 |
1 |
7.1 |
0 |
1.00e+00 |
Klhdc8b
Zfand2b
|
|
Ambulatory time before self-admin
|
20.51 |
2 |
0 |
0 |
0 |
1.00e+00 |
C1h11orf16
Zfand2b
|
|
Total heroin consumption
|
18.89 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Area under the delay curve
|
31.15 |
1 |
1 |
7.1 |
0 |
1.00e+00 |
Zfand2b
|
|
Delay disc. indifference point, 0s delay
|
28.7 |
1 |
1 |
7.1 |
0 |
1.00e+00 |
Klhdc8b
|
|
Delay disc. indifference point, 16s delay
|
21.3 |
4 |
1 |
7.1 |
-1 |
1.31e-07 |
Cnpy3
Rrp36
Trerf1
Zfand2b
|
|
Delay disc. indifference point, 24s delay
|
27.43 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zfand2b
|
|
Delay disc. indifference point, 4s delay
|
41.82 |
1 |
1 |
7.1 |
0 |
1.00e+00 |
Klhdc8b
|
|
Delay disc. indifference point, 8s delay
|
74.24 |
1 |
1 |
7.1 |
0 |
1.00e+00 |
Zfand2b
|
|
Delay discount exponential model param
|
16.14 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zfand2b
|
|
Delay discount hyperbolic model param
|
15.89 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zfand2b
|
|
Fecal boli incidents, locomotor time 1
|
96.73 |
2 |
1 |
7.1 |
0 |
1.00e+00 |
Klhdc8b
Zfand2b
|
|
Time >=10cm from walls, locomotor time 1
|
79.04 |
1 |
1 |
7.1 |
0 |
1.00e+00 |
Zfand2b
|
|
Rest time, locomotor task time 1
|
20.01 |
2 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
Nucb2
|
|
Distance moved, locomotor task time 1
|
31.01 |
1 |
1 |
7.1 |
0 |
1.00e+00 |
Klhdc8b
|
|
Vertical activity count, locomotor time 1
|
73.36 |
2 |
2 |
14.3 |
0 |
1.00e+00 |
Klhdc8b
Zfand2b
|
|
Bouts of movement, locomotor time 2
|
21.88 |
2 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
Zfand2b
|
|
Total resting periods, locomotor time 2
|
28.25 |
2 |
1 |
7.1 |
0 |
1.00e+00 |
Klhdc8b
Zfand2b
|
|
Rest time, locomotor task time 2
|
35.13 |
2 |
2 |
14.3 |
0 |
1.00e+00 |
Klhdc8b
Zfand2b
|
|
Distance moved, locomotor task time 2
|
63.13 |
2 |
2 |
14.3 |
0 |
1.00e+00 |
Klhdc8b
Zfand2b
|
|
Vertical activity count, locomotor time 2
|
34.04 |
2 |
1 |
7.1 |
0 |
1.00e+00 |
Klhdc8b
Zfand2b
|
|
Weight adjusted by age
|
30.32 |
1 |
1 |
7.1 |
0 |
1.00e+00 |
Klhdc8b
|
|
Seeking ratio, delayed vs. immediate footshock
|
12.86 |
5 |
0 |
0 |
-0.99 |
1.78e-07 |
C1h11orf16
Ces2c
Fam204a
Grk5
Sfxn4
|
|
Locomotion in novel chamber post-restriction
|
13.43 |
2 |
0 |
0 |
0 |
1.00e+00 |
C1h11orf16
Zfand2b
|
|
Food seeking constrained by brief footshock
|
13.85 |
2 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
Zfand2b
|
|
Run reversals in cocaine runway, females
|
36.45 |
1 |
1 |
7.1 |
0 |
1.00e+00 |
Zfand2b
|
|
Latency to leave start box in cocaine runway
|
34.23 |
1 |
1 |
7.1 |
0 |
1.00e+00 |
Zfand2b
|
|
Latency to leave start box in cocaine runway, F
|
31.9 |
1 |
1 |
7.1 |
0 |
1.00e+00 |
Zfand2b
|
|
Latency to leave start box in cocaine runway, M
|
55.97 |
1 |
1 |
7.1 |
0 |
1.00e+00 |
C1h11orf16
|
|
Cd content in liver
|
20.17 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Fe content in liver
|
40.39 |
1 |
1 |
7.1 |
0 |
1.00e+00 |
Klhdc8b
|
|
Mg content in liver
|
15.79 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Mn content in liver
|
12.13 |
1 |
0 |
0 |
0 |
1.00e+00 |
C1h11orf16
|
|
Na content in liver
|
16.36 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Rb content in liver
|
51.88 |
1 |
1 |
7.1 |
0 |
1.00e+00 |
Klhdc8b
|
|
Sr content in liver
|
22.41 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Zn content in liver
|
14.97 |
1 |
0 |
0 |
0 |
1.00e+00 |
C1h11orf16
|