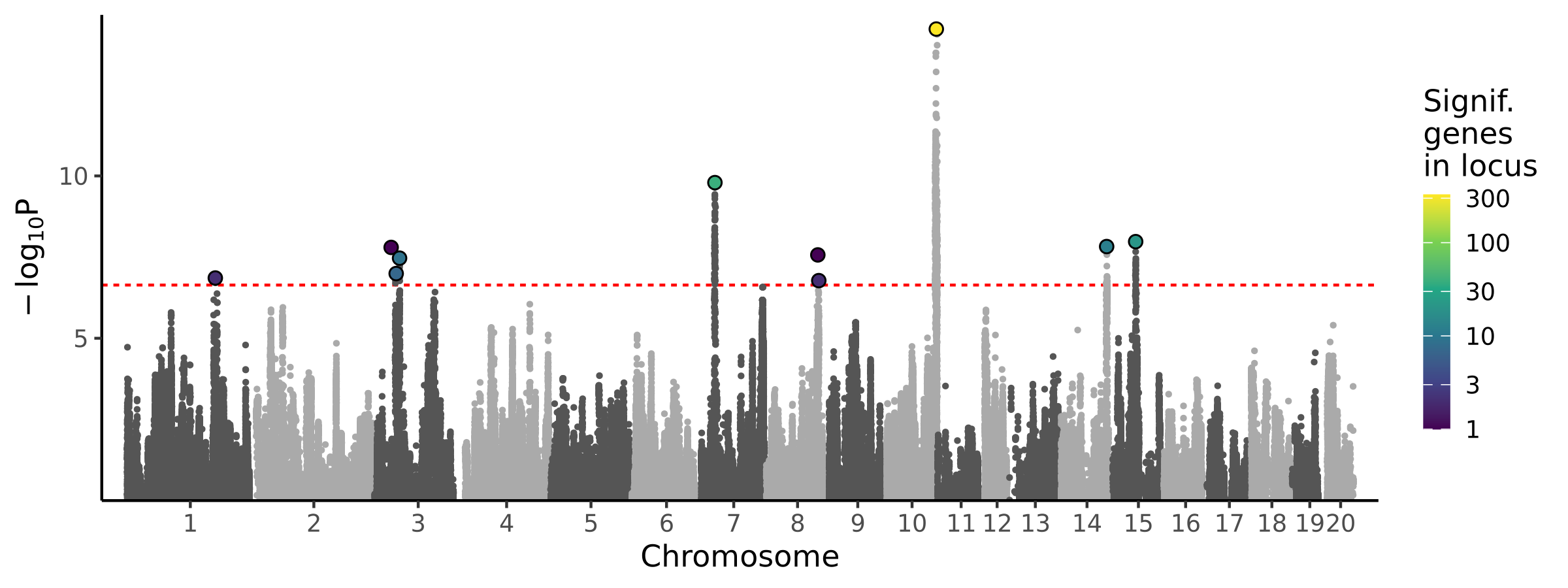
|
BMI with tail
|
643.95 |
5 |
1 |
2.7 |
-1 |
1.39e-08 |
Lta4h
Plxnc1
Prr5l
Retreg2
Vezt
|
|
BMI without tail
|
3352.73 |
15 |
2 |
5.4 |
0.94 |
6.22e-21 |
Cdk17
Cep83
Fgd6
LOC108351432
LOC134479863
LOC134481143
Lta4h
Metap2
Mir331
Ndufa12
Nr2c1
Plxnc1
Prr5l
Retreg2
Vezt
|
|
Body weight
|
9.32 |
60 |
26 |
70.3 |
0.63 |
3.27e-22 |
Aatk
Als2cl
Alyref
Amdhd1
B3gnt2
B3gntl1
Baiap2
Ccdc137
Ccdc38
Ccdc57
Cdk17
Cep131
Cep83
Cfap54
Eif4a3
Endov
Fasn
Fgd6
Gaa
Gcgr
Gps1
LOC102552414
LOC103693493
LOC108351432
LOC108352165
LOC120093559
LOC120095223
LOC120095311
LOC134479659
LOC134479863
LOC134480921
LOC134481143
Lrrfip2
Lta4h
Mcrip1
Metap2
Mir331
Mst1
Ndufa12
Nedd1
Nr2c1
Ntn4
Pcyt2
Pde6g
Plxnc1
Prr5l
Ptchd3
Retreg2
Rnf213
Rpl28l3
Sgsh
Slc16a3
Slc25a10
Slc38a10
Snrpf
Tmcc3
Tmem17
Tspan10
Vezt
Zfp950l6
|
|
Epididymis fat weight
|
54228.13 |
2 |
2 |
5.4 |
0 |
1.00e+00 |
Mst1
Retreg2
|
|
Glucose
|
623.51 |
4 |
2 |
5.4 |
-0.96 |
3.92e-02 |
B3gntl1
Fn3k
Mst1
Retreg2
|
|
Heart weight
|
13509 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Left kidney weight
|
1704.94 |
22 |
8 |
21.6 |
-0.86 |
4.63e-25 |
Amdhd1
Ccdc38
Cdk17
Cep83
Fgd6
LOC108351432
LOC134479863
LOC134481143
Lta4h
Metap2
Mir331
Ndufa12
Nedd1
Nr2c1
Ntn4
Plxnc1
Prr5l
Retreg2
Snrpf
Tmcc3
Tmem17
Vezt
|
|
Right kidney weight
|
1625.89 |
23 |
3 |
8.1 |
-0.87 |
2.79e-23 |
Amdhd1
B3gnt2
Ccdc38
Cdk17
Cep83
Fgd6
Lgalsl
LOC102552414
LOC108351432
LOC134479863
LOC134481143
Lta4h
Metap2
Mir331
Mst1
Ndufa12
Nr2c1
Ntn4
Plxnc1
Retreg2
Snrpf
Tmem17
Vezt
|
|
Tail length
|
370.35 |
61 |
20 |
54.1 |
0.81 |
9.57e-59 |
Aatk
Alyref
Amdhd1
Arhgdia
Baiap2
Card14
Cbx8
Ccdc137
Ccdc38
Ccdc57
Cd7
Cdk17
Cep131
Cep83
Cfap54
Chmp6
Eif4a3
Endov
Fgd6
Fscn2
Gaa
Gcgr
Hgs
LOC100912167
LOC103693493
LOC108351432
LOC108352165
LOC120093559
LOC120095223
LOC120095311
LOC134479659
LOC134479863
LOC134481143
Lta4h
Mcrip1
Metap2
Mir331
Ndufa12
Ndufaf8
Nedd1
Npb
Nploc4
Nptx1
Nr2c1
Ntn4
Oxld1
Pcyt2
Pde6g
Plxnc1
Retreg2
Rfng
Rnf213
Rptor
Sgsh
Slc16a3
Slc25a10
Slc38a10
Snrpf
Tmcc3
Tspan10
Vezt
|
|
Length with tail
|
61.69 |
45 |
26 |
70.3 |
0.98 |
1.94e-86 |
Aatk
Als2cl
Amdhd1
B3gnt2
Baiap2
Cbx8
Ccdc137
Ccdc38
Cdk17
Cep83
Cfap54
Commd1
Eif4a3
Endov
Fgd6
Gaa
LOC102551272
LOC102552414
LOC108351432
LOC108352165
LOC120093559
LOC134479659
LOC134479863
LOC134481143
Lrrfip2
Lta4h
Metap2
Mir331
Mst1
Ndufa12
Nedd1
Nptx1
Nr2c1
Ntn4
Plxnc1
Retreg2
Rnf213
Rptor
Sgsh
Slc38a10
Snrpf
Tbc1d16
Tmcc3
Tmem17
Vezt
|
|
Length without tail
|
1491.77 |
25 |
15 |
40.5 |
-0.84 |
1.68e-24 |
Als2cl
Amdhd1
Ccdc38
Cdk17
Cep83
Cfap54
Fgd6
LOC108351432
LOC120093559
LOC134479659
LOC134479863
LOC134481143
Lrrfip2
Lta4h
Metap2
Mir331
Mst1
Ndufa12
Nr2c1
Ntn4
Plxnc1
Retreg2
Snrpf
Tmcc3
Vezt
|
|
Liver weight, left
|
1824.75 |
4 |
2 |
5.4 |
-0.97 |
8.39e-05 |
Mst1
Ndufa12
Nr2c1
Retreg2
|
|
Liver weight, right
|
143.89 |
2 |
2 |
5.4 |
0 |
1.00e+00 |
Mst1
Retreg2
|
|
Parametrial fat weight
|
615.65 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Retroperitoneal fat weight
|
91.29 |
37 |
2 |
5.4 |
0.9 |
9.28e-69 |
Aatk
Alyref
Anapc11
Arhgdia
Baiap2
Card14
Ccdc137
Ccdc57
Cep131
Chmp6
Eif4a3
Endov
Fgd6
Gaa
Gcgr
LOC103693493
LOC108352165
LOC120095223
LOC120095311
Mcrip1
Mst1
Ndufaf8
Npb
Nploc4
Nr2c1
Oxld1
Pcyt2
Pde6g
Retreg2
Rfng
Rnf213
Rptor
Sgsh
Slc25a10
Slc38a10
Tepsin
Tspan10
|
|
Intraocular pressure
|
40071.91 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Extensor digitorum longus weight
|
166.06 |
9 |
1 |
2.7 |
0.9 |
5.28e-11 |
Cysltr2
Fndc3a
Itm2b
LOC134481143
Lpar6
Med4
Nudt15
Rcbtb2
Retreg2
|
|
Soleus weight
|
15.81 |
4 |
1 |
2.7 |
0.69 |
1.90e-02 |
Fndc3a
Nudt15
Rcbtb2
Retreg2
|
|
Tibialis anterior weight
|
1634.02 |
3 |
2 |
5.4 |
0 |
1.00e+00 |
LOC134481143
Prr5l
Retreg2
|
|
Number of licking bursts
|
157119.65 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Food consumed during 24 hour testing period
|
106196.53 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Times rat made contact with spout
|
305.36 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Mean time between licks in bursts
|
199507.71 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Mean num. licks in bursts
|
142600.29 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Std. dev. time between licks in bursts
|
94042.02 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Water consumed over 24 hours
|
103408.69 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Patch foraging indifference point 0 sec
|
115003.89 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Patch foraging indifference point AUC
|
44390.72 |
3 |
2 |
5.4 |
0 |
1.00e+00 |
Mst1
Prr5l
Retreg2
|
|
Indifference point function ln k
|
71040.92 |
2 |
2 |
5.4 |
0 |
1.00e+00 |
Mst1
Retreg2
|
|
Indifference point function log k
|
71040.92 |
2 |
2 |
5.4 |
0 |
1.00e+00 |
Mst1
Retreg2
|
|
Patch foraging total patch changes 0 sec
|
120264.14 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Patch foraging total patch changes 12 sec
|
3637.8 |
3 |
2 |
5.4 |
0 |
1.00e+00 |
Mst1
Prr5l
Retreg2
|
|
Patch foraging total patch changes 18 sec
|
58889.67 |
2 |
1 |
2.7 |
0 |
1.00e+00 |
Mst1
Retreg2
|
|
Patch foraging total patch changes 24 sec
|
83554.6 |
2 |
2 |
5.4 |
0 |
1.00e+00 |
Mst1
Retreg2
|
|
Patch foraging total patch changes 6 sec
|
16325.11 |
2 |
1 |
2.7 |
0 |
1.00e+00 |
Prr5l
Retreg2
|
|
Patch foraging time to switch 0 sec
|
78318.27 |
2 |
2 |
5.4 |
0 |
1.00e+00 |
Prr5l
Retreg2
|
|
Patch foraging water rate 0 sec
|
74407.85 |
2 |
2 |
5.4 |
0 |
1.00e+00 |
Prr5l
Retreg2
|
|
Patch foraging water rate 12 sec
|
161858.08 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Patch foraging water rate 18 sec
|
51645.28 |
2 |
1 |
2.7 |
0 |
1.00e+00 |
Prr5l
Retreg2
|
|
Patch foraging water rate 24 sec
|
4691.77 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Patch foraging water rate 6 sec
|
3954.86 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Locomotor activity
|
15.72 |
2 |
1 |
2.7 |
0 |
1.00e+00 |
Mst1
Retreg2
|
|
Locomotor testing distance
|
44732.22 |
2 |
2 |
5.4 |
0 |
1.00e+00 |
Mst1
Retreg2
|
|
Locomotor testing rearing
|
76214.5 |
2 |
2 |
5.4 |
0 |
1.00e+00 |
Mst1
Retreg2
|
|
Light reinforcement 1
|
155242.72 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Light reinforcement 2
|
53965.57 |
2 |
2 |
5.4 |
0 |
1.00e+00 |
Mst1
Retreg2
|
|
Reaction time number correct
|
153465.22 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Reaction time mean minus median
|
205810.62 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Reaction time mean minus median AUC
|
104201.52 |
2 |
2 |
5.4 |
0 |
1.00e+00 |
Mst1
Retreg2
|
|
Reaction time num false alarms
|
28542.61 |
2 |
1 |
2.7 |
0 |
1.00e+00 |
Mst1
Retreg2
|
|
Reaction time num false alarms AUC
|
106496.51 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Reaction time trials correct on left
|
153465.22 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Reaction time trials on left
|
149991.58 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Reaction time mean
|
94380.53 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Reaction time mean AUC
|
113341.21 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Median of all reaction times
|
31417.12 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Reaction time omissions
|
34267.91 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Reaction time false alarm rate
|
891.26 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Reaction time premature initiation rate
|
36877.02 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Reaction time premature initiations
|
138718.63 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Std. dev. reaction times
|
74746.42 |
2 |
1 |
2.7 |
0 |
1.00e+00 |
Mst1
Retreg2
|
|
Reaction time trials completed
|
149991.58 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Reaction time trials AUC
|
151696.42 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Social response proportion
|
17657.99 |
2 |
1 |
2.7 |
0 |
1.00e+00 |
Mst1
Retreg2
|
|
Social responses
|
1905.06 |
3 |
1 |
2.7 |
0 |
1.00e+00 |
Mst1
Prr5l
Retreg2
|
|
Social time
|
22125.12 |
2 |
1 |
2.7 |
0 |
1.00e+00 |
Prr5l
Retreg2
|
|
Conditioned locomotion
|
6074.35 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Cocaine response after cond. corrected
|
65812.39 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Cocaine response after cond. not corrected
|
64247.35 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Cocaine response before conditioning
|
2800.41 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Saline control response
|
6522.43 |
2 |
2 |
5.4 |
0 |
1.00e+00 |
Mst1
Retreg2
|
|
Condit. Reinf. active minus inactive responses
|
26712.34 |
2 |
1 |
2.7 |
0 |
1.00e+00 |
Mst1
Retreg2
|
|
Condit. Reinf. active-inactive response ratio
|
91582.24 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Condit. Reinf. active responses
|
4856.3 |
2 |
2 |
5.4 |
0 |
1.00e+00 |
Mst1
Retreg2
|
|
Condit. Reinf. inactive responses
|
21067.54 |
2 |
2 |
5.4 |
0 |
1.00e+00 |
Mst1
Retreg2
|
|
Condit. Reinf. lever presses
|
7960.29 |
2 |
2 |
5.4 |
0 |
1.00e+00 |
Mst1
Retreg2
|
|
Condit. Reinf. lever reinforcers received
|
1007.39 |
2 |
1 |
2.7 |
0 |
1.00e+00 |
Mst1
Retreg2
|
|
Pavlov. Cond. lever latency
|
3648.25 |
3 |
1 |
2.7 |
0 |
1.00e+00 |
Mst1
Prr5l
Retreg2
|
|
Pavlov. Cond. magazine entry latency
|
24773.4 |
2 |
2 |
5.4 |
0 |
1.00e+00 |
Mst1
Retreg2
|
|
Pavlov. Cond. change in total contacts
|
114781.94 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Pavlov. Cond. index score
|
24017.04 |
2 |
2 |
5.4 |
0 |
1.00e+00 |
Mst1
Retreg2
|
|
Pavlov. Cond. latency score
|
29259.45 |
2 |
2 |
5.4 |
0 |
1.00e+00 |
Mst1
Retreg2
|
|
Pavlov. Cond. lever contacts
|
7372.88 |
3 |
2 |
5.4 |
0 |
1.00e+00 |
Mst1
Prr5l
Retreg2
|
|
Pavlov. Cond. magazine entry number
|
8453.95 |
2 |
2 |
5.4 |
0 |
1.00e+00 |
Mst1
Retreg2
|
|
Pavlov. Cond. intertrial magazine entries
|
52501.71 |
2 |
2 |
5.4 |
0 |
1.00e+00 |
Mst1
Retreg2
|
|
Pavlov. Cond. lever-magazine prob. diff.
|
22461.1 |
2 |
2 |
5.4 |
0 |
1.00e+00 |
Mst1
Retreg2
|
|
Pavlov. Cond. response bias
|
2629.8 |
2 |
2 |
5.4 |
0 |
1.00e+00 |
Mst1
Retreg2
|
|
Conditioned reinforcement - actives
|
191492.54 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Conditioned reinforcement - inactives
|
77543.68 |
2 |
2 |
5.4 |
0 |
1.00e+00 |
Mst1
Retreg2
|
|
Conditioned locomotion
|
41346.85 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Intermittent access intake day 1-15 change
|
105304.97 |
2 |
1 |
2.7 |
0 |
1.00e+00 |
Mst1
Retreg2
|
|
Intermittent access intake escalation
|
104773.43 |
2 |
1 |
2.7 |
0 |
1.00e+00 |
Mst1
Retreg2
|
|
Intermittent access intake escalation 2
|
107995.31 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Intermitt. access day 1 inactive lever presses
|
118279.42 |
2 |
2 |
5.4 |
0 |
1.00e+00 |
Mst1
Retreg2
|
|
Intermitt. access day 15 inactive lever presses
|
208241.66 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Intermitt. access escalation Index
|
92516.01 |
2 |
2 |
5.4 |
0 |
1.00e+00 |
Mst1
Retreg2
|
|
Intermittent access day 1 total infusions
|
155525.68 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Intermittent access day 15 total infusions
|
131485.34 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Intermittent access terminal intake (last 3 days)
|
83205.14 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Intermittent access total infusions
|
675.9 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Intermittent access day 1 locomotion
|
104459.41 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Intermittent access day 15 total locomotion
|
177836.92 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Intermittent access total locomotion
|
711.59 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Intermittent access standard deviation
|
100956.84 |
2 |
2 |
5.4 |
0 |
1.00e+00 |
Mst1
Retreg2
|
|
Cocaine induced anxiety
|
160280.46 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Post-drug Anxiety
|
20460.83 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Baseline Anxiety
|
103471.3 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Lifetime Intake
|
6830.03 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Incentive Sensitization - Responses
|
163579.25 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Incentive Sensitization - Breakpoint
|
142790.8 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Progressive ratio test 1 active lever presses
|
99872.69 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Progressive ratio test 1 breakpoint
|
96247.61 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Progressive ratio test 1 inactive lever presses
|
75556.75 |
2 |
2 |
5.4 |
0 |
1.00e+00 |
Mst1
Retreg2
|
|
Progressive ratio test 2 active lever presses
|
57222.6 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Progressive ratio test 2 breakpoint
|
34975.96 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Progressive ratio test 2 inactive lever presses
|
164104.71 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Total sessions with >9 infusions
|
3995.08 |
2 |
2 |
5.4 |
0 |
1.00e+00 |
Mst1
Retreg2
|
|
Short access day 1 total inactive lever presses
|
38149.28 |
3 |
2 |
5.4 |
0 |
1.00e+00 |
Mst1
Prr5l
Retreg2
|
|
Short access day 10 total inactive lever presses
|
108578.43 |
2 |
2 |
5.4 |
0 |
1.00e+00 |
Mst1
Retreg2
|
|
Short access day 1 total infusions
|
111479.43 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Short access day 10 total infusions
|
38157.63 |
2 |
2 |
5.4 |
0 |
1.00e+00 |
Mst1
Retreg2
|
|
Short access total infusions
|
33946.03 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Short access day 1 locomotion
|
42032.93 |
2 |
1 |
2.7 |
0 |
1.00e+00 |
Prr5l
Retreg2
|
|
Short access day 10 total locomotion
|
90801.14 |
2 |
1 |
2.7 |
0 |
1.00e+00 |
Mst1
Retreg2
|
|
Short access total locomotion
|
22766.31 |
3 |
2 |
5.4 |
0 |
1.00e+00 |
Mst1
Prr5l
Retreg2
|
|
Compulsive drug intake
|
65439.45 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
One hour access (0.3 mA shock)
|
9669.44 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
One hour access (shock baseline)
|
140263.4 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Number of responses in last shaping day
|
25656.23 |
2 |
2 |
5.4 |
0 |
1.00e+00 |
Prr5l
Retreg2
|
|
Context. condit. distance diff. score
|
970.59 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Locomotion velocity, session 1
|
14615.25 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Locomotion distance, session 1
|
4416.18 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Locomotion velocity, session 2
|
4702.2 |
2 |
2 |
5.4 |
0 |
1.00e+00 |
Mst1
Retreg2
|
|
Locomotion distance, session 2
|
2393.66 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Locomotion velocity, session 3
|
1177.39 |
2 |
1 |
2.7 |
0 |
1.00e+00 |
Prr5l
Retreg2
|
|
Locomotion distance, session 3
|
16615.78 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Stereotopy head waving bouts, day 3
|
1968.48 |
3 |
1 |
2.7 |
0 |
1.00e+00 |
Mst1
Prr5l
Retreg2
|
|
Stereotopy head waving duration, day 3
|
6074.03 |
2 |
1 |
2.7 |
0 |
1.00e+00 |
Mst1
Retreg2
|
|
Locomotion velocity, session 7
|
708.6 |
2 |
2 |
5.4 |
0 |
1.00e+00 |
Mst1
Retreg2
|
|
Locomotion distance, session 7
|
1934.91 |
2 |
2 |
5.4 |
0 |
1.00e+00 |
Mst1
Retreg2
|
|
Stereotopy head waving bouts, day 7
|
10466.92 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Stereotopy head waving duration, day 7
|
8847.36 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Locomotion velocity, session 8
|
1892.98 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Locomotion distance, session 8
|
673.32 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Degree of sensitization distance
|
7696.03 |
2 |
2 |
5.4 |
0 |
1.00e+00 |
Mst1
Retreg2
|
|
Degree of sensitization stereotypy
|
8496.38 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Condit. Reinf. active minus inactive responses
|
2077.92 |
2 |
2 |
5.4 |
0 |
1.00e+00 |
Prr5l
Retreg2
|
|
Condit. Reinf. active-inactive response ratio
|
558.58 |
2 |
1 |
2.7 |
0 |
1.00e+00 |
Prr5l
Retreg2
|
|
Condit. Reinf. active responses
|
5847.95 |
2 |
2 |
5.4 |
0 |
1.00e+00 |
Prr5l
Retreg2
|
|
Condit. Reinf. inactive responses
|
19375.3 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Incentive salience index mean
|
4947.1 |
3 |
2 |
5.4 |
0 |
1.00e+00 |
Mst1
Prr5l
Retreg2
|
|
Condit. Reinf. lever presses
|
7168.4 |
3 |
3 |
8.1 |
0 |
1.00e+00 |
Mst1
Prr5l
Retreg2
|
|
Time in familiar zone, hab. session 1
|
41873.03 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Time in novel zone, hab. session 1
|
11855.76 |
3 |
2 |
5.4 |
0 |
1.00e+00 |
Mst1
Prr5l
Retreg2
|
|
Total zone transitions, hab. session 1
|
1818.53 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Total locomotion distance, hab. session 1
|
1687 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Locomotion velocity, hab. session 1
|
1131.06 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Time in familiar zone, hab. session 2
|
11707.23 |
2 |
2 |
5.4 |
0 |
1.00e+00 |
Mst1
Retreg2
|
|
Time in novel zone, hab. session 2
|
12499.19 |
2 |
2 |
5.4 |
0 |
1.00e+00 |
Mst1
Retreg2
|
|
Total zone transitions, hab. session 2
|
280.53 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Total locomotion distance, hab. session 2
|
55.8 |
2 |
1 |
2.7 |
0 |
1.00e+00 |
Prr5l
Retreg2
|
|
Locomotion velocity, hab. session 2
|
79.6 |
2 |
1 |
2.7 |
0 |
1.00e+00 |
Prr5l
Retreg2
|
|
Novel to familiar place preference ratio
|
18514.75 |
3 |
1 |
2.7 |
0 |
1.00e+00 |
Mst1
Prr5l
Retreg2
|
|
Novelty place preference
|
18140.34 |
3 |
1 |
2.7 |
0 |
1.00e+00 |
Mst1
Prr5l
Retreg2
|
|
Time in novel zone, NPP test
|
13626.92 |
3 |
1 |
2.7 |
0 |
1.00e+00 |
Mst1
Prr5l
Retreg2
|
|
Total zone transitions, NPP test
|
8230.03 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Total locomotion distance, NPP test
|
7114 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Locomotion velocity, NPP test
|
4041.22 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Pavlov. Cond. lever latency
|
1120.34 |
2 |
2 |
5.4 |
0 |
1.00e+00 |
Mst1
Retreg2
|
|
Pavlov. Cond. change in total contacts
|
2281.88 |
2 |
1 |
2.7 |
0 |
1.00e+00 |
Prr5l
Retreg2
|
|
Pavlov. Cond. index score
|
899.58 |
2 |
2 |
5.4 |
0 |
1.00e+00 |
Mst1
Retreg2
|
|
Pavlov. Cond. latency score
|
628.33 |
3 |
2 |
5.4 |
0 |
1.00e+00 |
Mst1
Prr5l
Retreg2
|
|
Pavlov. Cond. lever contacts
|
1539.83 |
2 |
2 |
5.4 |
0 |
1.00e+00 |
Mst1
Retreg2
|
|
Pavlov. Cond. magazine entry number
|
1093.11 |
2 |
2 |
5.4 |
0 |
1.00e+00 |
Prr5l
Retreg2
|
|
Pavlov. Cond. intertrial magazine entries
|
198.69 |
3 |
3 |
8.1 |
0 |
1.00e+00 |
Mst1
Prr5l
Retreg2
|
|
Pavlov. Cond. lever-magazine prob. diff.
|
979.4 |
2 |
1 |
2.7 |
0 |
1.00e+00 |
Mst1
Retreg2
|
|
Pavlov. Cond. response bias
|
880.36 |
3 |
1 |
2.7 |
0 |
1.00e+00 |
Mst1
Prr5l
Retreg2
|
|
Bone: apparent density
|
22268.03 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Bone surface
|
8383.75 |
3 |
2 |
5.4 |
0 |
1.00e+00 |
Mst1
Prr5l
Retreg2
|
|
Bone volume
|
33831.7 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Bone: connectivity density
|
3566.21 |
2 |
2 |
5.4 |
0 |
1.00e+00 |
Prr5l
Retreg2
|
|
Bone: cortical apparent density
|
45266.32 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Bone: cortical area
|
1472 |
8 |
3 |
8.1 |
0.99 |
5.71e-11 |
B3gntl1
Ccdc57
Fn3k
Mst1
Rab40b
Retreg2
Slc16a3
Zfp950l6
|
|
Bone: cortical porosity
|
932.97 |
2 |
2 |
5.4 |
0 |
1.00e+00 |
Prr5l
Retreg2
|
|
Bone: cortical porosity
|
2417.92 |
2 |
2 |
5.4 |
0 |
1.00e+00 |
Prr5l
Retreg2
|
|
Bone: cortical thickness
|
73.67 |
15 |
6 |
16.2 |
-0.7 |
3.52e-05 |
Afmid
Ccdc57
Cd7
Cenpx
Engase
Fasn
Mst1
Rab40b
Rbfox3
Retreg2
Slc16a3
Timp2
Usp36
Uts2r
Zfp950l6
|
|
Bone: cortical thickness
|
6116.53 |
2 |
2 |
5.4 |
0 |
1.00e+00 |
Mst1
Retreg2
|
|
Bone: cortical tissue density
|
48453.42 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Bone: elastic displacement
|
29626.25 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Bone: elastic work
|
84.3 |
16 |
3 |
8.1 |
-0.77 |
5.29e-05 |
Ccdc137
Ccdc57
Cd7
Cenpx
Fasn
Gps1
Lrrc45
Mcrip1
Mst1
Narf
Rab40b
Retreg2
Slc16a3
Slc38a10
Tepsin
Uts2r
|
|
Bone: endosteal estimation
|
35257.61 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Bone: endosteal perimeter
|
2986.84 |
9 |
2 |
5.4 |
0.98 |
4.61e-11 |
Afmid
Birc5
C1qtnf1
Dnah17
Lgals3bp
LOC102548046
Pgs1
Retreg2
Tmem235
|
|
Bone: final force
|
483.1 |
9 |
3 |
8.1 |
1 |
5.05e-15 |
B3gntl1
Ccdc57
Fn3k
Mst1
Ptchd3
Rab40b
Retreg2
Slc16a3
Zfp950l6
|
|
Bone: final moment
|
45.39 |
17 |
3 |
8.1 |
0.98 |
2.30e-21 |
B3gntl1
Ccdc57
Cenpx
Csnk1d
Dcxr
Fasn
Fn3k
Gps1
LOC134480921
Mcrip1
Mst1
Ptchd3
Rab40b
Retreg2
Slc16a3
Tbcd
Zfp950l6
|
|
Bone: marrow area
|
37067.95 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Bone: maximum diameter
|
287.21 |
18 |
2 |
5.4 |
0.94 |
2.05e-43 |
Afmid
Birc5
C1qtnf1
Cyth1
Dnah17
Lgals3bp
LOC100912319
LOC102548046
LOC120095218
Mst1
Pgs1
Retreg2
Syngr2
Tha1
Tk1
Tmc6
Tmem235
Tnrc6c
|
|
Bone: maximum force
|
1451.52 |
13 |
4 |
10.8 |
0.98 |
6.18e-16 |
B3gntl1
Ccdc57
Fasn
Fn3k
Gps1
LOC134480921
Mst1
Ptchd3
Rab40b
Retreg2
Slc16a3
Tbcd
Zfp950l6
|
|
Bone: maximum moment
|
629.27 |
18 |
5 |
13.5 |
0.97 |
3.40e-24 |
B3gntl1
Ccdc57
Cenpx
Csnk1d
Dcxr
Fasn
Fn3k
Gps1
LOC134480921
Metrnl
Mst1
Ptchd3
Rab40b
Retreg2
Slc16a3
Tbcd
Zfp950l11
Zfp950l6
|
|
Bone: minimum diameter
|
8606.13 |
2 |
2 |
5.4 |
0 |
1.00e+00 |
Mst1
Retreg2
|
|
Bone: periosteal estimation
|
9560.62 |
4 |
2 |
5.4 |
1 |
4.31e-03 |
B3gntl1
Mst1
Retreg2
Zfp950l6
|
|
Bone: periosteal perimeter
|
1401.96 |
15 |
2 |
5.4 |
0.97 |
2.77e-19 |
Afmid
Birc5
Cyth1
Dnah17
LOC120095218
Mst1
Pgs1
Rab40b
Retreg2
Syngr2
Tha1
Tmc6
Tmem235
Tnrc6c
Zfp950l6
|
|
Bone: post-yield work
|
50847.58 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Bone: stiffness
|
17808.98 |
2 |
2 |
5.4 |
0 |
1.00e+00 |
Mst1
Retreg2
|
|
Bone: tissue strength
|
12264.67 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Bone: trabecular number
|
41138.39 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Bone: trabecular spacing
|
15656.57 |
2 |
2 |
5.4 |
0 |
1.00e+00 |
Mst1
Retreg2
|
|
Bone: trabecular thickness
|
16647.3 |
2 |
2 |
5.4 |
0 |
1.00e+00 |
Mst1
Retreg2
|
|
Bone: trabecular tissue density
|
17883.86 |
3 |
3 |
8.1 |
0 |
1.00e+00 |
Mst1
Prr5l
Retreg2
|
|
Breakpoint value in progressive ratio session
|
112428.06 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Distance traveled before self-admin
|
134013.73 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Distance traveled after self-admin
|
100820.36 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Delta time in closed arm before/after self-admin
|
1271.86 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Delta time in open arm before/after self-admin
|
54811.83 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Diff in mean of infusions in LGA sessions
|
54252.87 |
2 |
1 |
2.7 |
0 |
1.00e+00 |
Mst1
Retreg2
|
|
Extinction: sum of active levers before priming
|
45807.86 |
2 |
2 |
5.4 |
0 |
1.00e+00 |
Mst1
Retreg2
|
|
Active lever presses in extinction session 6
|
1571.88 |
2 |
1 |
2.7 |
0 |
1.00e+00 |
Mst1
Retreg2
|
|
Delta distance traveled before/after self-admin
|
12434.77 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Delta ambulatory time before/after self-admin
|
96034.01 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Time in closed arm before self-admin
|
44133.6 |
2 |
2 |
5.4 |
0 |
1.00e+00 |
Mst1
Retreg2
|
|
Time in closed arm after self-admin
|
9759.54 |
2 |
2 |
5.4 |
0 |
1.00e+00 |
Mst1
Retreg2
|
|
Time in open arm before self-admin
|
34587.99 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Time in open arm after self-admin
|
979.73 |
2 |
2 |
5.4 |
0 |
1.00e+00 |
Mst1
Retreg2
|
|
Sum of active levers in priming session hrs 5-6
|
3877.89 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Time to tail flick, vehicle, before self-admin
|
25677.46 |
2 |
1 |
2.7 |
0 |
1.00e+00 |
Mst1
Retreg2
|
|
Time to tail flick, vehicle, after self-admin
|
118286.85 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Time to tail flick, test, before self-admin
|
32803.11 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Time to tail flick, test, after self-admin
|
5828.87 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Delta time to tail flick, vehicle, before/after SA
|
18281.67 |
2 |
2 |
5.4 |
0 |
1.00e+00 |
Mst1
Retreg2
|
|
Delta time to tail flick, test, before/after SA
|
56832.34 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Ambulatory time before self-admin
|
171079.71 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Ambulatory time after self-admin
|
88230.17 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Total heroin consumption
|
961.5 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Area under the delay curve
|
81605.23 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Delay disc. indifference point, 0s delay
|
9477.47 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Delay disc. indifference point, 16s delay
|
74803.61 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Delay disc. indifference point, 2s delay
|
825.84 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Delay disc. indifference point, 24s delay
|
99983.64 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Delay disc. indifference point, 4s delay
|
2525.7 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Delay disc. indifference point, 8s delay
|
101423.99 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Delay discount exponential model param
|
60214.95 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Delay discount hyperbolic model param
|
45715.54 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Fecal boli incidents, locomotor time 1
|
150160.89 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Change in fecal boli incidents, locomotor task
|
23481.3 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Time >=10cm from walls, locomotor time 1
|
49255.61 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Bouts of movement, locomotor time 1
|
41590.88 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Total resting periods, locomotor time 1
|
11802.64 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Rest time, locomotor task time 1
|
75951.88 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Distance moved, locomotor task time 1
|
159283.67 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Vertical activity count, locomotor time 1
|
219061.96 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Time >=10cm from walls, locomotor time 2
|
161986.9 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Bouts of movement, locomotor time 2
|
174298.32 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Total resting periods, locomotor time 2
|
190079.87 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Rest time, locomotor task time 2
|
83778.18 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Distance moved, locomotor task time 2
|
34268.85 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Vertical activity count, locomotor time 2
|
138414.05 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Weight adjusted by age
|
161973.18 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Seeking ratio, delayed vs. immediate footshock
|
49395.56 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Locomotion in novel chamber
|
8159.66 |
2 |
1 |
2.7 |
0 |
1.00e+00 |
Prr5l
Retreg2
|
|
Locomotion in novel chamber post-restriction
|
154642.04 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Progressive ratio
|
157894.89 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Food seeking constrained by brief footshock
|
444.28 |
8 |
1 |
2.7 |
-1 |
1.22e-19 |
Cysltr2
Fndc3a
Itm2b
Lpar6
Med4
Nudt15
Rcbtb2
Retreg2
|
|
Seeking ratio, punishment vs. effort
|
118281.2 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Run reversals in cocaine runway, females
|
37174.37 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Run reversals in cocaine runway, males
|
29861.56 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Latency to leave start box in cocaine runway
|
58575.8 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Latency to leave start box in cocaine runway, F
|
28379.24 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Latency to leave start box in cocaine runway, M
|
7140.1 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Co content in liver
|
2368.81 |
2 |
1 |
2.7 |
0 |
1.00e+00 |
Prr5l
Retreg2
|
|
Cu content in liver
|
162.43 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Fe content in liver
|
5656.2 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
K content in liver
|
6593.53 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Mg content in liver
|
24493.99 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Mn content in liver
|
4374.36 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Na content in liver
|
16641.94 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Rb content in liver
|
428.45 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|
|
Zn content in liver
|
15755.62 |
1 |
1 |
2.7 |
0 |
1.00e+00 |
Retreg2
|