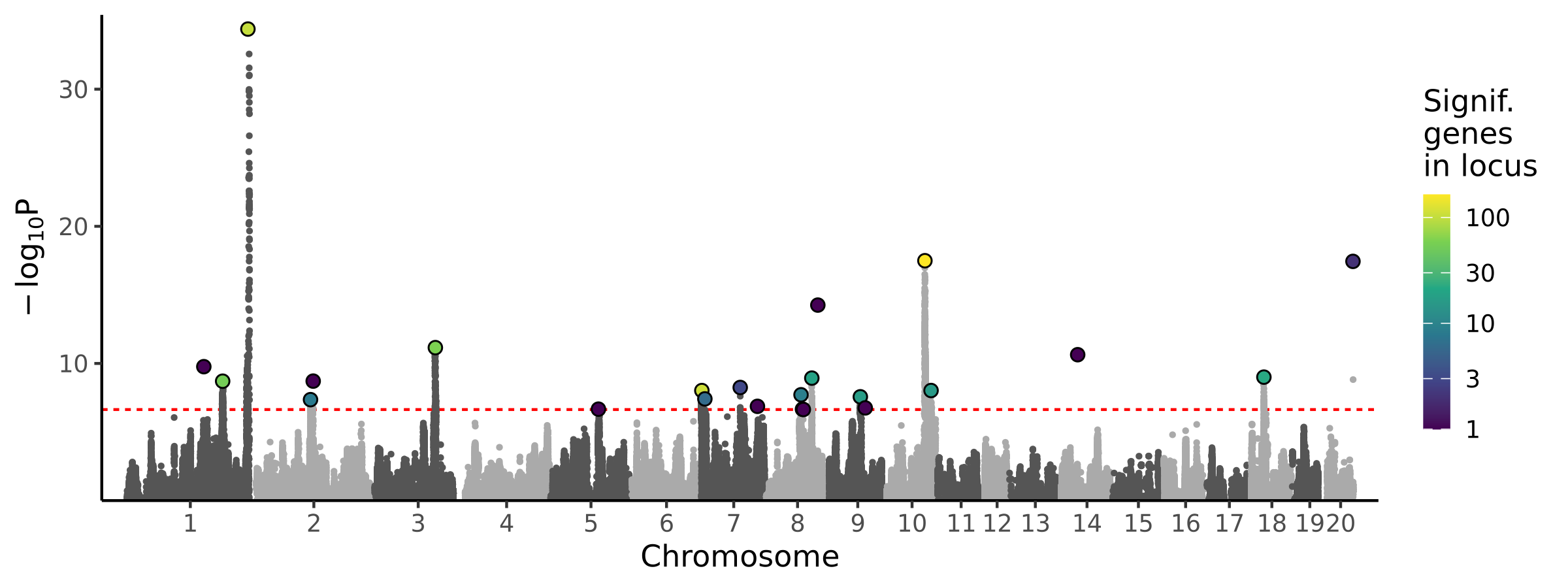
|
BMI without tail
|
15.05 |
35 |
18 |
35.3 |
0.98 |
3.90e-126 |
Arsg
C1h11orf16
Cacul1
Cep112
Ces2c
Dennd10
Dstn
Emx2os
Epb41l4a
Fam204a
Grk5
Hspa12a
Kif16b
Klhdc8b
LOC102548590
LOC102550729
LOC120100068
LOC120101717
LOC120101719
Nanos1
Otor
Pcsk2
Pls1
Prdx3
Prlhr
Rab11fip2
Rrbp1
Sec23b
Sfxn4
Snrpb2
Trpc1
Trps1
U2surp
Zfand2b
Zfp950
|
|
Body weight
|
13.06 |
115 |
90 |
176.5 |
0.99 |
0.00e+00 |
Ablim1
Acsl5
Afap1l2
Amz2
Apc
Apoh
Arsg
Atp11b
Atp1b3
Atrnl1
Axin2
Brd8
Cacul1
Casp7
Ccdc186
Ccpg1
Cd99
Cdc23
Cep112
Ces2c
Cox8a
Dclre1a
Dennd10
Dnaaf4
Dstn
Dtd1
Dzank1
Ehbp1l1
Emx2os
Eno4
Epb41l4a
Fam13b
Fam204a
Fermt3
Fhip2a
Flrt1
Gclc
Gfra1
Gpam
Grk5
Gucy2g
Habp2
Hmgcll1
Hspa12a
Jade1
Kcnk4
Kif16b
Kif20a
Klhdc8b
LOC102547573
LOC102548590
LOC102550112
LOC102550304
LOC102550729
LOC103694460
LOC120095197
LOC120095201
LOC120100065
LOC120100068
LOC120101717
LOC120101719
Macrod1
Map2k6
Mccc1
Naaladl1
Nanos1
Nhlrc2
Nme5
Nrap
Otor
Otub1
Ovol1
PCOLCE2
Pcsk2
Pigb
Pkd2l2
Plekhs1
Pls1
Polr3f
Prdx3
Prlhr
Pygo1
Rab11fip2
Rbbp9
Reep5
Rpl37-ps2
Rps6ka4
Rrbp1
Rtn3
Sac3d1
Sec23b
Sfxn4
Slc16a6
Smim26
Snrpb2
Snx15
Snx32
Srp19
Syvn1
Tafa5
Tbkbp1
Tcf7l2
Tectb
Tfdp2
Trpc1
Trps1
Trub1
U2surp
Wipi1
Xrn1
Zdhhc6
Zfp133
Zfp267
Zfp950
Znrd2
|
|
Epididymis fat weight
|
19.4 |
59 |
45 |
88.2 |
0.98 |
6.46e-222 |
Ablim1
Acsl5
Afap1l2
Amz2
Apoh
Arsg
Atrnl1
Axin2
Cacul1
Casp7
Ccdc186
Cep112
Ces2c
Dclre1a
Dennd10
Dstn
Emx2os
Eno4
Fam204a
Fhip2a
Gfra1
Gpam
Grk5
Gucy2g
Habp2
Hspa12a
Kif16b
LOC102547573
LOC102550112
LOC102550304
LOC102550729
LOC120095201
LOC120100065
LOC120100068
LOC120101717
LOC120101719
Nanos1
Nhlrc2
Nrap
Otor
Pcsk2
Plekhs1
Prdx3
Prlhr
Rab11fip2
Rbbp9
Rrbp1
Sec23b
Sfxn4
Slc16a6
Snrpb2
Tcf7l2
Tectb
Trub1
Wipi1
Zdhhc6
Zfand2b
Zfp133
Zfp950
|
|
Heart weight
|
20.2 |
5 |
1 |
2 |
0.86 |
6.54e-03 |
Cacul1
Emx2os
Klhdc8b
Sfxn4
Zfand2b
|
|
Left kidney weight
|
11.64 |
24 |
18 |
35.3 |
0.98 |
1.34e-65 |
Acsl5
Cacul1
Cd99
Ces2c
Dennd10
Emx2os
Fam204a
Gpam
Grk5
Gucy2g
Klhdc8b
LOC102550729
LOC103694460
LOC120100068
Nanos1
Ociad2
Prdx3
Prlhr
Rab11fip2
Sfxn4
Tafa5
Trpc1
Zfand2b
Zfp950
|
|
Right kidney weight
|
10.32 |
37 |
18 |
35.3 |
0.97 |
1.79e-82 |
Ablim1
Acsl5
Afap1l2
Atrnl1
Cacul1
Casp7
Ccdc186
Cd99
Ces2c
Dclre1a
Dennd10
Emx2os
Fam204a
Fhip2a
Gpam
Grk5
Gucy2g
Habp2
Klhdc8b
LOC102550729
LOC103694460
LOC120100068
Nanos1
Nhlrc2
Nrap
Ociad2
Plekhs1
Prdx3
Prlhr
Rab11fip2
Sfxn4
Tafa5
Tectb
Trub1
U2surp
Zfand2b
Zfp950
|
|
Tail length
|
11.63 |
61 |
39 |
76.5 |
-0.99 |
0.00e+00 |
Abi3
Apof
Atp5mc1
B4galnt2
Calcoco2
Cbx1
Cdk5rap3
Cnpy2
Copz2
Cryga
Crygc
Dgka
Dnajc14
Erbb3
Gip
Gls2
Gngt2
Hoxb2
Hoxb3
Hoxb4
Hoxb7
Hoxb9
Hsd17b6
Il23a
Klhdc8b
Kpnb1
LOC102546598
LOC102547047
LOC102547278
LOC102547955
LOC102550580
LOC108352124
LOC120093525
LOC120095179
LOC120095180
Lrrc46
Mrpl10
Myl6
Nfe2l1
Osbpl7
Phb1
Phospho1
Pnpo
Prim1
Prr15l
Ptges3
Rbms2
Rnf41
Sarnp
Scrn2
Skap1
Smarcc2
Snf8
Snx11
Sp2
Spop
Stat2
Timeless
Ube2z
Zfand2b
Zfp652
|
|
Length with tail
|
7.25 |
56 |
35 |
68.6 |
-0.93 |
1.66e-174 |
Abi3
Atp5mc1
B4galnt2
Cacul1
Calcoco2
Cbx1
Ccnyl1
Cdk5rap3
Copz2
Cryga
Crygc
Fam237a
Fzd5
Gip
Gngt2
Hoxb2
Hoxb3
Hoxb4
Hoxb7
Hoxb9
Idh1
Kif16b
Klhdc8b
Kpnb1
LOC102546598
LOC102547047
LOC102547278
LOC102547955
LOC102550580
LOC108352124
LOC120094695
LOC120095179
LOC120095180
Lrrc46
Mettl21a
Mrpl10
Nfe2l1
Osbpl7
Otor
Phb1
Phospho1
Pikfyve
Plekhm3
Pnpo
Prr15l
Scrn2
Sfxn4
Skap1
Snf8
Snrpb2
Snx11
Sp2
Spop
Ube2z
Zfand2b
Zfp652
|
|
Length without tail
|
13.13 |
8 |
1 |
2 |
0.83 |
2.34e-04 |
Acsl5
Cacul1
Casp7
Emx2os
Grk5
Habp2
Prlhr
Zfand2b
|
|
Liver weight, left
|
11.57 |
6 |
0 |
0 |
0.54 |
2.14e-01 |
C1h11orf16
Cacul1
Grk5
Klhdc8b
LOC103694460
Prlhr
|
|
Liver weight, right
|
13.27 |
18 |
7 |
13.7 |
0.95 |
1.05e-25 |
Acsl5
Cacul1
Casp7
Cd99
Dennd10
Fam204a
Gpam
Grk5
Habp2
LOC103694460
Nanos1
Plekhs1
Prlhr
Rab11fip2
Sfxn4
Zdhhc6
Zfand2b
Zfp950
|
|
Parametrial fat weight
|
17.16 |
53 |
30 |
58.8 |
0.99 |
2.67e-252 |
Ablim1
Acsl5
Afap1l2
Atrnl1
Cacul1
Casp7
Ccdc186
Ces2c
Dclre1a
Dennd10
Dstn
Dtd1
Dzank1
Emx2os
Eno4
Fam204a
Fhip2a
Gfra1
Gpam
Grk5
Gucy2g
Habp2
Hspa12a
Kif16b
LOC102547573
LOC102550304
LOC102550729
LOC120100065
LOC120100068
LOC120101717
LOC120101719
Nanos1
Nhlrc2
Nrap
Otor
Pcsk2
Plekhs1
Polr3f
Prdx3
Prlhr
Rab11fip2
Rbbp9
Rrbp1
Sec23b
Sfxn4
Smim26
Snrpb2
Tcf7l2
Trub1
Zdhhc6
Zfand2b
Zfp133
Zfp950
|
|
Retroperitoneal fat weight
|
22.54 |
66 |
55 |
107.8 |
0.99 |
0.00e+00 |
Ablim1
Acsl5
Afap1l2
Amz2
Apoh
Arsg
Atrnl1
Axin2
Cacul1
Casp7
Ccdc186
Cd99
Cep112
Ces2c
Dclre1a
Dennd10
Dstn
Dtd1
Dzank1
Emx2os
Eno4
Fam204a
Fhip2a
Gfra1
Gpam
Grk5
Gucy2g
Habp2
Hspa12a
Kif16b
Klhdc8b
LOC102547573
LOC102550112
LOC102550304
LOC102550729
LOC103694460
LOC120095201
LOC120100065
LOC120100068
LOC120101717
LOC120101719
Nanos1
Nhlrc2
Nrap
Otor
Pcsk2
Plekhs1
Polr3f
Prdx3
Prlhr
Rab11fip2
Rbbp9
Rrbp1
Sec23b
Sfxn4
Slc16a6
Smim26
Snrpb2
Tcf7l2
Tectb
Trub1
Wipi1
Zdhhc6
Zfand2b
Zfp133
Zfp950
|
|
Intraocular pressure
|
38.04 |
2 |
2 |
3.9 |
0 |
1.00e+00 |
LOC103694460
Zfand2b
|
|
Extensor digitorum longus weight
|
5.76 |
71 |
11 |
21.6 |
1 |
4.97e-263 |
Apof
Atp11b
Atp5f1b
Baz2a
Bloc1s1
Ccpg1
Cdk2
Cnpy2
Coq10a
Cox8a
Cs
Dgka
Dnaaf4
Dnajc14
Ehbp1l1
Erbb3
Fermt3
Gls2
Hsd17b6
Ikzf4
Itga7
Jade1
Kcnk4
LOC102546625
LOC102549817
LOC102551925
LOC102556486
LOC103692757
LOC103694460
LOC108350028
LOC120093525
LOC120093754
LOC130046242
Macrod1
Mccc1
Mfsd12
Myl6
Nabp2
Naca
Ormdl2
Otub1
Ovol1
Pan2
Pcnx3
Pigb
Prim1
Ptges3
Pygo1
Rbms2
Rdh5
RGD1562465
Rnf41
Rpl37-ps2
Rps6ka4
Rtn3
Sac3d1
Sarnp
Sirt6
Smarcc2
Snx15
Snx32
Stat2
Suox
Syvn1
Tbkbp1
Tfdp2
Timeless
Tmem198b
Zc3h10
Zfp267
Znrd2
|
|
Soleus weight
|
7.3 |
55 |
8 |
15.7 |
0.99 |
3.76e-170 |
Ablim1
Acsl5
Apof
Atrnl1
Bloc1s1
Ccpg1
Cdk2
Cnpy2
Coq10a
Cs
Dgka
Dnaaf4
Dnajc14
Emx2os
Erbb3
Gls2
Gpam
Grk5
Gucy2g
Habp2
Hsd17b6
Ikzf4
Itga7
Klhdc8b
LOC102546625
LOC102549817
LOC102551925
LOC103692757
LOC120093525
LOC120093754
LOC120100068
LOC130046242
Matk
Mfsd12
Myl6
Nabp2
Ormdl2
Pan2
Pigb
Prim1
Ptges3
Pygo1
Rbms2
Rdh5
Rnf41
Sarnp
Sirt6
Smarcc2
Stat2
Suox
Tectb
Timeless
Tmem198b
Trub1
Zc3h10
|
|
Tibialis anterior weight
|
8.25 |
22 |
9 |
17.6 |
0.99 |
1.33e-35 |
Atp11b
Ccpg1
Cox8a
Dnaaf4
Fermt3
Gclc
Jade1
Kcnk4
LOC103694460
Macrod1
Mccc1
Otub1
Pigb
Pygo1
Rps6ka4
Rtn3
Sac3d1
Snx15
Snx32
Syvn1
Zfp267
Znrd2
|
|
Tibia length
|
7.6 |
6 |
1 |
2 |
1 |
9.52e-18 |
Kif16b
Klhdc8b
LOC120101717
Ociad2
Otor
Snrpb2
|
|
Number of licking bursts
|
28.51 |
2 |
2 |
3.9 |
0 |
1.00e+00 |
Klhdc8b
Zfand2b
|
|
Food consumed during 24 hour testing period
|
15.78 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zfand2b
|
|
Mean num. licks in bursts
|
18.13 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zfand2b
|
|
Water consumed over 24 hours
|
34.33 |
1 |
1 |
2 |
0 |
1.00e+00 |
Zfand2b
|
|
Patch foraging indifference point 0 sec
|
12.38 |
2 |
0 |
0 |
0 |
1.00e+00 |
LOC103694460
Zfand2b
|
|
Patch foraging indifference point AUC
|
24.2 |
1 |
1 |
2 |
0 |
1.00e+00 |
Zfand2b
|
|
Indifference point function ln k
|
35.82 |
3 |
2 |
3.9 |
0 |
1.00e+00 |
Klhdc8b
Ociad2
Zfand2b
|
|
Indifference point function log k
|
35.82 |
3 |
2 |
3.9 |
0 |
1.00e+00 |
Klhdc8b
Ociad2
Zfand2b
|
|
Patch foraging total patch changes 0 sec
|
15.52 |
3 |
1 |
2 |
0 |
1.00e+00 |
LOC103694460
Prlhr
Zfand2b
|
|
Patch foraging total patch changes 18 sec
|
30.97 |
2 |
1 |
2 |
0 |
1.00e+00 |
Ociad2
Zfand2b
|
|
Patch foraging total patch changes 24 sec
|
45.89 |
2 |
1 |
2 |
0 |
1.00e+00 |
Ociad2
Zfand2b
|
|
Patch foraging total patch changes 6 sec
|
16.36 |
3 |
1 |
2 |
0 |
1.00e+00 |
Fhip2a
LOC120102905
Zfand2b
|
|
Patch foraging time to switch 0 sec
|
33.47 |
1 |
1 |
2 |
0 |
1.00e+00 |
Zfand2b
|
|
Patch foraging water rate 0 sec
|
39.47 |
1 |
1 |
2 |
0 |
1.00e+00 |
Zfand2b
|
|
Patch foraging water rate 12 sec
|
17.89 |
8 |
1 |
2 |
-0.53 |
1.17e-01 |
Ehbp1l1
Ovol1
Rps6ka4
Sac3d1
Snx15
Snx32
Zfand2b
Znrd2
|
|
Patch foraging water rate 24 sec
|
12.61 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zfand2b
|
|
Patch foraging water rate 6 sec
|
35.43 |
1 |
1 |
2 |
0 |
1.00e+00 |
Zfand2b
|
|
Locomotor activity
|
35.08 |
2 |
1 |
2 |
0 |
1.00e+00 |
Klhdc8b
Zfand2b
|
|
Locomotor testing distance
|
55.86 |
2 |
2 |
3.9 |
0 |
1.00e+00 |
Klhdc8b
Zfand2b
|
|
Locomotor testing rearing
|
99.51 |
1 |
1 |
2 |
0 |
1.00e+00 |
Zfand2b
|
|
Light reinforcement 1
|
19.25 |
1 |
1 |
2 |
0 |
1.00e+00 |
Zfand2b
|
|
Reaction time number correct
|
66.72 |
2 |
2 |
3.9 |
0 |
1.00e+00 |
Klhdc8b
Zfand2b
|
|
Reaction time mean minus median
|
98.07 |
2 |
1 |
2 |
0 |
1.00e+00 |
C1h11orf16
Zfand2b
|
|
Reaction time mean minus median AUC
|
122.84 |
2 |
1 |
2 |
0 |
1.00e+00 |
C1h11orf16
Zfand2b
|
|
Reaction time num false alarms
|
17.2 |
1 |
0 |
0 |
0 |
1.00e+00 |
Ociad2
|
|
Reaction time num false alarms AUC
|
27.43 |
1 |
1 |
2 |
0 |
1.00e+00 |
Zfand2b
|
|
Reaction time trials correct on left
|
66.72 |
2 |
2 |
3.9 |
0 |
1.00e+00 |
Klhdc8b
Zfand2b
|
|
Reaction time trials on left
|
63.1 |
2 |
2 |
3.9 |
0 |
1.00e+00 |
Klhdc8b
Zfand2b
|
|
Reaction time mean
|
36.11 |
1 |
1 |
2 |
0 |
1.00e+00 |
Zfand2b
|
|
Reaction time mean AUC
|
33.75 |
1 |
1 |
2 |
0 |
1.00e+00 |
Zfand2b
|
|
Median of all reaction times
|
17.65 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zfand2b
|
|
Reaction time omissions
|
27.35 |
4 |
3 |
5.9 |
-0.66 |
3.44e-01 |
Jade1
Klhdc8b
Ociad2
Zfand2b
|
|
Reaction time false alarm rate
|
20.24 |
1 |
1 |
2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Reaction time premature initiations
|
41.6 |
1 |
1 |
2 |
0 |
1.00e+00 |
Zfand2b
|
|
Std. dev. reaction times
|
43.56 |
2 |
1 |
2 |
0 |
1.00e+00 |
Jade1
Zfand2b
|
|
Reaction time trials completed
|
63.1 |
2 |
2 |
3.9 |
0 |
1.00e+00 |
Klhdc8b
Zfand2b
|
|
Reaction time trials AUC
|
64.7 |
2 |
2 |
3.9 |
0 |
1.00e+00 |
Klhdc8b
Zfand2b
|
|
Conditioned locomotion
|
28.4 |
1 |
1 |
2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Cocaine response after cond. not corrected
|
16.6 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Cocaine response before conditioning
|
38.66 |
1 |
1 |
2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Condit. Reinf. active responses
|
14.98 |
4 |
0 |
0 |
0.88 |
1.23e-01 |
Epb41l4a
Fam13b
Ociad2
Reep5
|
|
Condit. Reinf. lever reinforcers received
|
13.58 |
3 |
0 |
0 |
0 |
1.00e+00 |
Fam13b
Ociad2
Reep5
|
|
Pavlov. Cond. index score
|
15.43 |
1 |
0 |
0 |
0 |
1.00e+00 |
LOC103694460
|
|
Pavlov. Cond. magazine entry number
|
25.13 |
2 |
1 |
2 |
0 |
1.00e+00 |
Klhdc8b
LOC103694460
|
|
Pavlov. Cond. response bias
|
13.13 |
1 |
0 |
0 |
0 |
1.00e+00 |
LOC103694460
|
|
Conditioned reinforcement - actives
|
152.54 |
1 |
1 |
2 |
0 |
1.00e+00 |
Zfand2b
|
|
Conditioned reinforcement - inactives
|
76.2 |
2 |
1 |
2 |
0 |
1.00e+00 |
Klhdc8b
Zfand2b
|
|
Intermittent access intake day 1-15 change
|
156.77 |
1 |
1 |
2 |
0 |
1.00e+00 |
Zfand2b
|
|
Intermittent access intake escalation
|
117.41 |
1 |
1 |
2 |
0 |
1.00e+00 |
Zfand2b
|
|
Intermittent access intake escalation 2
|
17.78 |
1 |
0 |
0 |
0 |
1.00e+00 |
C1h11orf16
|
|
Intermitt. access day 1 inactive lever presses
|
169.63 |
1 |
1 |
2 |
0 |
1.00e+00 |
Zfand2b
|
|
Intermitt. access day 15 inactive lever presses
|
162.52 |
2 |
2 |
3.9 |
0 |
1.00e+00 |
Klhdc8b
Zfand2b
|
|
Intermitt. access escalation Index
|
38.02 |
1 |
1 |
2 |
0 |
1.00e+00 |
Ociad2
|
|
Intermittent access day 1 total infusions
|
71.42 |
1 |
1 |
2 |
0 |
1.00e+00 |
Zfand2b
|
|
Intermittent access day 15 total infusions
|
35.39 |
1 |
1 |
2 |
0 |
1.00e+00 |
Zfand2b
|
|
Intermittent access terminal intake (last 3 days)
|
24.92 |
2 |
1 |
2 |
0 |
1.00e+00 |
Klhdc8b
Zfand2b
|
|
Intermittent access total infusions
|
15.45 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Intermittent access day 1 locomotion
|
26.47 |
1 |
1 |
2 |
0 |
1.00e+00 |
Zfand2b
|
|
Intermittent access day 15 total locomotion
|
93.08 |
1 |
1 |
2 |
0 |
1.00e+00 |
Zfand2b
|
|
Intermittent access standard deviation
|
229.66 |
2 |
1 |
2 |
0 |
1.00e+00 |
Klhdc8b
Zfand2b
|
|
Cocaine induced anxiety
|
125.35 |
1 |
1 |
2 |
0 |
1.00e+00 |
Zfand2b
|
|
Post-drug Anxiety
|
21.14 |
1 |
0 |
0 |
0 |
1.00e+00 |
Ociad2
|
|
Baseline Anxiety
|
393.92 |
1 |
1 |
2 |
0 |
1.00e+00 |
Zfand2b
|
|
Incentive Sensitization - Responses
|
309.83 |
1 |
1 |
2 |
0 |
1.00e+00 |
Zfand2b
|
|
Incentive Sensitization - Breakpoint
|
300.45 |
1 |
1 |
2 |
0 |
1.00e+00 |
Zfand2b
|
|
Progressive ratio test 1 active lever presses
|
26.41 |
1 |
1 |
2 |
0 |
1.00e+00 |
Zfand2b
|
|
Progressive ratio test 1 breakpoint
|
19.19 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zfand2b
|
|
Progressive ratio test 1 inactive lever presses
|
16.55 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Progressive ratio test 2 active lever presses
|
109.22 |
1 |
1 |
2 |
0 |
1.00e+00 |
Zfand2b
|
|
Progressive ratio test 2 breakpoint
|
106.65 |
1 |
1 |
2 |
0 |
1.00e+00 |
Zfand2b
|
|
Progressive ratio test 2 inactive lever presses
|
63.47 |
1 |
1 |
2 |
0 |
1.00e+00 |
Zfand2b
|
|
Total sessions with >9 infusions
|
16 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Short access day 1 total inactive lever presses
|
21.93 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zfand2b
|
|
Short access day 10 total inactive lever presses
|
72.35 |
2 |
2 |
3.9 |
0 |
1.00e+00 |
Klhdc8b
Zfand2b
|
|
Short access day 1 locomotion
|
129.19 |
1 |
1 |
2 |
0 |
1.00e+00 |
Zfand2b
|
|
Short access day 10 total locomotion
|
432.47 |
1 |
1 |
2 |
0 |
1.00e+00 |
Zfand2b
|
|
Short access total locomotion
|
209.96 |
1 |
1 |
2 |
0 |
1.00e+00 |
Zfand2b
|
|
Compulsive drug intake
|
68.39 |
1 |
1 |
2 |
0 |
1.00e+00 |
Zfand2b
|
|
One hour access (0.3 mA shock)
|
22.58 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zfand2b
|
|
Number of responses in last shaping day
|
67.75 |
1 |
1 |
2 |
0 |
1.00e+00 |
Zfand2b
|
|
Locomotion distance, session 1
|
23.98 |
1 |
1 |
2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Locomotion distance, session 2
|
33.15 |
1 |
1 |
2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Locomotion velocity, session 3
|
17.14 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Locomotion distance, session 8
|
52.78 |
1 |
1 |
2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Degree of sensitization distance
|
24.75 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Condit. Reinf. active minus inactive responses
|
35.59 |
1 |
1 |
2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Condit. Reinf. active-inactive response ratio
|
25.6 |
1 |
1 |
2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Condit. Reinf. active responses
|
18.23 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Incentive salience index mean
|
26.23 |
1 |
1 |
2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Time in familiar zone, hab. session 1
|
63.49 |
1 |
1 |
2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Total zone transitions, hab. session 1
|
37.38 |
1 |
1 |
2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Time in familiar zone, hab. session 2
|
27.1 |
1 |
1 |
2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Total zone transitions, hab. session 2
|
69.58 |
1 |
1 |
2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Total locomotion distance, hab. session 2
|
19.2 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Locomotion velocity, hab. session 2
|
21.15 |
1 |
1 |
2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Novel to familiar place preference ratio
|
22.33 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Novelty place preference
|
20.99 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Time in novel zone, NPP test
|
17.06 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Total zone transitions, NPP test
|
23.4 |
5 |
1 |
2 |
0.94 |
1.97e-04 |
Klhdc8b
PCOLCE2
Pls1
Trpc1
U2surp
|
|
Total locomotion distance, NPP test
|
47.64 |
1 |
1 |
2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Locomotion velocity, NPP test
|
51.12 |
1 |
1 |
2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Pavlov. Cond. change in total contacts
|
69.48 |
1 |
1 |
2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Pavlov. Cond. index score
|
14.07 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Pavlov. Cond. lever-magazine prob. diff.
|
14.82 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Bone volume
|
10.02 |
2 |
0 |
0 |
0 |
1.00e+00 |
Cacul1
Grk5
|
|
Bone: cortical apparent density
|
15.5 |
1 |
0 |
0 |
0 |
1.00e+00 |
C1h11orf16
|
|
Bone: cortical area
|
13.31 |
26 |
0 |
0 |
0.97 |
4.57e-63 |
Ablim1
Acsl5
Afap1l2
Cacul1
Casp7
Ccdc186
Ces2c
Dclre1a
Dennd10
Emx2os
Fam204a
Fhip2a
Gfra1
Grk5
Habp2
LOC102547573
LOC102550729
LOC120100065
LOC120100068
Nhlrc2
Plekhs1
Prlhr
Rab11fip2
Sfxn4
Trub1
Zfp950
|
|
Bone: cortical porosity
|
16.96 |
1 |
0 |
0 |
0 |
1.00e+00 |
LOC103694460
|
|
Bone: cortical thickness
|
11.62 |
6 |
0 |
0 |
0.93 |
1.06e-04 |
Emx2os
Fam204a
LOC120100068
Prlhr
Rab11fip2
Zfp950
|
|
Bone: cortical tissue density
|
16.13 |
1 |
0 |
0 |
0 |
1.00e+00 |
C1h11orf16
|
|
Bone: endosteal perimeter
|
14.86 |
3 |
1 |
2 |
0 |
1.00e+00 |
Ablim1
Cd99
LOC103694460
|
|
Bone: final force
|
11.01 |
7 |
0 |
0 |
0.99 |
3.11e-14 |
Cacul1
Emx2os
Grk5
Prlhr
Rab11fip2
Sfxn4
Zfp950
|
|
Bone: final moment
|
11.67 |
6 |
0 |
0 |
1 |
5.48e-18 |
Cacul1
Grk5
Prlhr
Rab11fip2
Sfxn4
Zfp950
|
|
Bone: maximum diameter
|
10.95 |
18 |
1 |
2 |
0.99 |
4.04e-40 |
Ablim1
Acsl5
Afap1l2
Cacul1
Casp7
Ccdc186
Cd99
Dclre1a
Fhip2a
Gfra1
Habp2
LOC102547573
LOC103694460
LOC120100065
Nhlrc2
Nrap
Plekhs1
Trub1
|
|
Bone: maximum force
|
10.64 |
6 |
0 |
0 |
1 |
1.62e-11 |
Cacul1
Grk5
Prlhr
Rab11fip2
Sfxn4
Zfp950
|
|
Bone: maximum moment
|
11.11 |
6 |
0 |
0 |
1 |
3.40e-14 |
Cacul1
Grk5
Prlhr
Rab11fip2
Sfxn4
Zfp950
|
|
Bone: minimum diameter
|
9.23 |
3 |
0 |
0 |
0 |
1.00e+00 |
Cacul1
LOC103694460
Ociad2
|
|
Bone: periosteal estimation
|
9.12 |
16 |
0 |
0 |
0.98 |
8.89e-34 |
Acsl5
Cacul1
Casp7
Ces2c
Dennd10
Emx2os
Fam204a
Grk5
Habp2
LOC120100065
Nhlrc2
Plekhs1
Prlhr
Rab11fip2
Sfxn4
Zfp950
|
|
Bone: periosteal perimeter
|
13.14 |
27 |
3 |
5.9 |
0.95 |
1.58e-65 |
Ablim1
Acsl5
Afap1l2
Atrnl1
Cacul1
Casp7
Ccdc186
Ces2c
Dclre1a
Dennd10
Fam204a
Fhip2a
Gfra1
Grk5
Habp2
LOC102547573
LOC102550729
LOC103694460
LOC120100065
Nhlrc2
Nrap
Plekhs1
Prlhr
Rab11fip2
Sfxn4
Trub1
Zfp950
|
|
Bone: post-yield work
|
13.79 |
9 |
0 |
0 |
1 |
8.89e-24 |
Arl2
Capn1
Fosl1
Naaladl1
Sac3d1
Scyl1
Sipa1
Snx15
Snx32
|
|
Bone: stiffness
|
13.41 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Bone: trabecular number
|
11.4 |
1 |
0 |
0 |
0 |
1.00e+00 |
C1h11orf16
|
|
Bone: trabecular spacing
|
15.64 |
1 |
0 |
0 |
0 |
1.00e+00 |
C1h11orf16
|
|
Bone: trabecular thickness
|
15.7 |
5 |
0 |
0 |
1 |
1.06e-12 |
Cacul1
Grk5
Prlhr
Sfxn4
Zfp950
|
|
Distance traveled before self-admin
|
17.44 |
1 |
0 |
0 |
0 |
1.00e+00 |
C1h11orf16
|
|
Delta time in open arm before/after self-admin
|
26.67 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zfand2b
|
|
Active lever presses in extinction session 6
|
26.41 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zfand2b
|
|
Time in closed arm before self-admin
|
44.32 |
1 |
1 |
2 |
0 |
1.00e+00 |
Zfand2b
|
|
Time in open arm before self-admin
|
24.67 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zfand2b
|
|
Time to tail flick, vehicle, after self-admin
|
56.6 |
2 |
2 |
3.9 |
0 |
1.00e+00 |
Klhdc8b
Zfand2b
|
|
Delta time to tail flick, vehicle, before/after SA
|
74.6 |
1 |
1 |
2 |
0 |
1.00e+00 |
Zfand2b
|
|
Ambulatory time before self-admin
|
25.36 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zfand2b
|
|
Total heroin consumption
|
18.89 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Area under the delay curve
|
31.15 |
1 |
1 |
2 |
0 |
1.00e+00 |
Zfand2b
|
|
Delay disc. indifference point, 0s delay
|
28.7 |
1 |
1 |
2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Delay disc. indifference point, 16s delay
|
69.03 |
1 |
1 |
2 |
0 |
1.00e+00 |
Zfand2b
|
|
Delay disc. indifference point, 24s delay
|
27.43 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zfand2b
|
|
Delay disc. indifference point, 4s delay
|
41.82 |
1 |
1 |
2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Delay disc. indifference point, 8s delay
|
74.24 |
1 |
1 |
2 |
0 |
1.00e+00 |
Zfand2b
|
|
Delay discount exponential model param
|
16.14 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zfand2b
|
|
Delay discount hyperbolic model param
|
15.89 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zfand2b
|
|
Fecal boli incidents, locomotor time 1
|
96.73 |
2 |
1 |
2 |
0 |
1.00e+00 |
Klhdc8b
Zfand2b
|
|
Time >=10cm from walls, locomotor time 1
|
79.04 |
1 |
1 |
2 |
0 |
1.00e+00 |
Zfand2b
|
|
Rest time, locomotor task time 1
|
22.55 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Distance moved, locomotor task time 1
|
31.01 |
1 |
1 |
2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Vertical activity count, locomotor time 1
|
73.36 |
2 |
2 |
3.9 |
0 |
1.00e+00 |
Klhdc8b
Zfand2b
|
|
Bouts of movement, locomotor time 2
|
21.88 |
2 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
Zfand2b
|
|
Total resting periods, locomotor time 2
|
28.25 |
2 |
1 |
2 |
0 |
1.00e+00 |
Klhdc8b
Zfand2b
|
|
Rest time, locomotor task time 2
|
35.13 |
2 |
2 |
3.9 |
0 |
1.00e+00 |
Klhdc8b
Zfand2b
|
|
Distance moved, locomotor task time 2
|
63.13 |
2 |
2 |
3.9 |
0 |
1.00e+00 |
Klhdc8b
Zfand2b
|
|
Vertical activity count, locomotor time 2
|
54.92 |
1 |
1 |
2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Weight adjusted by age
|
30.32 |
1 |
1 |
2 |
0 |
1.00e+00 |
Klhdc8b
|
|
Seeking ratio, delayed vs. immediate footshock
|
18.76 |
1 |
0 |
0 |
0 |
1.00e+00 |
C1h11orf16
|
|
Locomotion in novel chamber post-restriction
|
14.04 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zfand2b
|
|
Food seeking constrained by brief footshock
|
15.76 |
2 |
0 |
0 |
0 |
1.00e+00 |
LOC120102905
Zfand2b
|
|
Run reversals in cocaine runway, females
|
36.45 |
1 |
1 |
2 |
0 |
1.00e+00 |
Zfand2b
|
|
Latency to leave start box in cocaine runway
|
34.23 |
1 |
1 |
2 |
0 |
1.00e+00 |
Zfand2b
|
|
Latency to leave start box in cocaine runway, F
|
31.9 |
1 |
1 |
2 |
0 |
1.00e+00 |
Zfand2b
|
|
Latency to leave start box in cocaine runway, M
|
55.97 |
1 |
1 |
2 |
0 |
1.00e+00 |
C1h11orf16
|
|
Cd content in liver
|
20.17 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Cu content in liver
|
23.31 |
4 |
1 |
2 |
-0.99 |
7.22e-03 |
Cd99
LOC103694460
LOC120095179
Snf8
|
|
Fe content in liver
|
33.68 |
2 |
2 |
3.9 |
0 |
1.00e+00 |
Klhdc8b
LOC103694460
|
|
K content in liver
|
34.63 |
2 |
1 |
2 |
0 |
1.00e+00 |
Cd99
LOC103694460
|
|
Na content in liver
|
16.36 |
1 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
|
|
Rb content in liver
|
54.46 |
3 |
3 |
5.9 |
0 |
1.00e+00 |
Cd99
Klhdc8b
LOC103694460
|
|
Se content in liver
|
41.97 |
1 |
1 |
2 |
0 |
1.00e+00 |
Ociad2
|
|
Sr content in liver
|
21.09 |
3 |
0 |
0 |
0 |
1.00e+00 |
Klhdc8b
LOC103694460
Ociad2
|