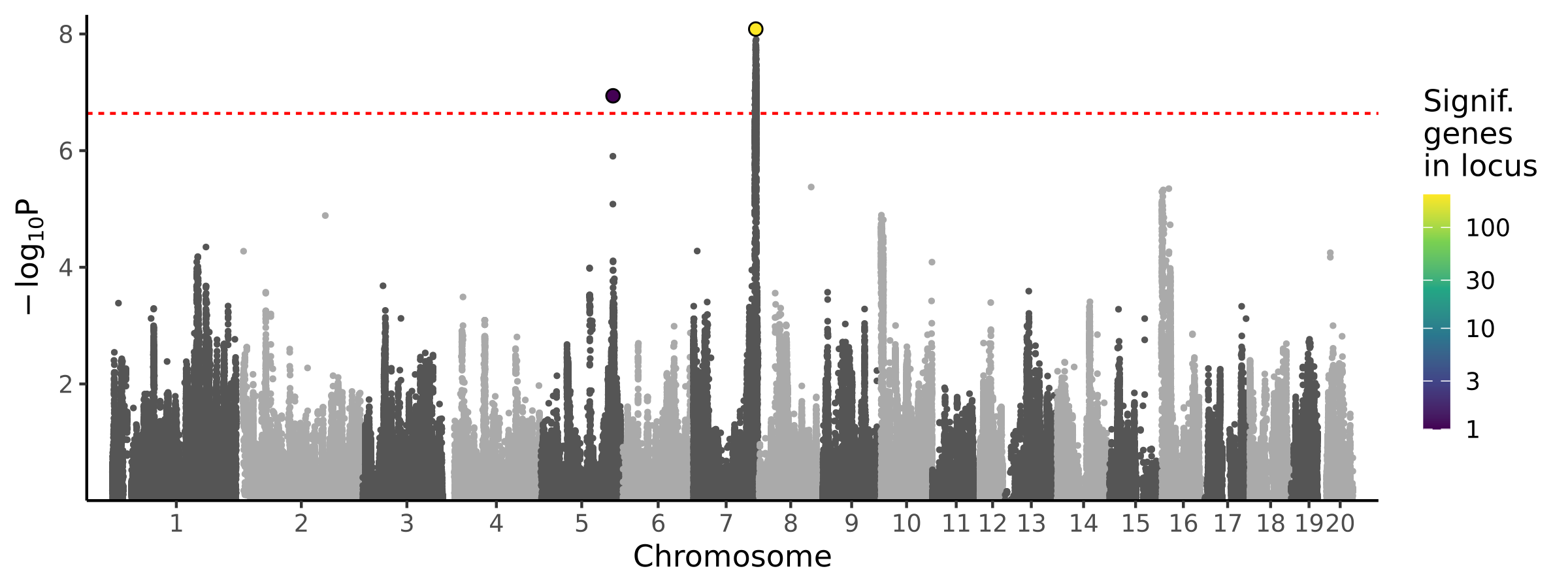
|
BMI with tail
|
5.04 |
42 |
0 |
0 |
1 |
1.23e-185 |
Adcy6
Aqp5
Asb8
Asic1
Atf1
Bcdin3d
Cacnb3
Ccdc65
Ccnt1
Cela1
Ddx23
Dhh
Dnajc22
Faim2
Fkbp11
Kansl2
Letmd1
Lmbr1l
LOC100362820
LOC102546778
LOC108351553
LOC108351554
LOC120093735
LOC120093830
Mettl7a
Nckap5l
Olr1877
Pfkm
Prkag1
Prph
Rhebl1
Rnd1
Slc11a2
Slc48a1
Smagp
Smarcd1
Spats2
Tmbim6
Tuba1a
Tuba1b
Tuba1c
Zfp641
|
|
Body weight
|
5.76 |
70 |
32 |
457.1 |
1 |
0.00e+00 |
Adcy6
Aqp5
Asb8
Asic1
Atf1
Bcdin3d
Bin2
C1ql4
Cacnb3
Ccdc65
Ccnt1
Cela1
Cers5
Cox14
Csrnp2
Ddn
Ddx23
Dhh
Dip2b
Dnajc22
Faim2
Fkbp11
Fmnl3
Gpd1
Kansl2
Kcnh3
Kmt2d
Larp4
Letmd1
Lima1
Lmbr1l
LOC100362820
LOC102546778
LOC102548155
LOC102555574
LOC102556302
LOC103692978
LOC108351553
LOC108351554
LOC120093732
LOC120093735
LOC120093830
LOC120093831
LOC120093832
Mcrs1
Mettl7a
Nckap5l
Olr1877
Pfkm
Prkag1
Prpf40b
Prph
Rhebl1
Rnd1
Senp1
Slc11a2
Slc48a1
Slc4a8
Smagp
Smarcd1
Spats2
Tmbim6
Tmco4
Tmprss12
Tuba1a
Tuba1b
Tuba1c
Wnt1
Wnt10b
Zfp641
|
|
Length without tail
|
7.64 |
45 |
0 |
0 |
1 |
0.00e+00 |
Adcy6
Asb8
Asic1
Bcdin3d
C1ql4
Cacnb3
Ccdc65
Ccnt1
Ddn
Ddx23
Dhh
Dnajc22
Faim2
Fkbp11
Fmnl3
Kansl2
Kcnh3
Kmt2d
Lmbr1l
LOC100362820
LOC102546778
LOC102548155
LOC102555574
LOC102556302
LOC103692978
LOC120093732
LOC120093830
LOC120093831
Mcrs1
Nckap5l
Olr1877
Prkag1
Prpf40b
Prph
Rhebl1
Rnd1
Senp1
Slc48a1
Spats2
Tmbim6
Tuba1a
Tuba1b
Tuba1c
Wnt1
Wnt10b
|
|
Extensor digitorum longus weight
|
4.74 |
61 |
0 |
0 |
1 |
0.00e+00 |
Adcy6
Aqp5
Asb8
Asic1
Atf1
Bcdin3d
Bin2
Cacnb3
Ccdc65
Ccnt1
Cela1
Cers5
Cox14
Ddx23
Dhh
Dip2b
Dnajc22
Faim2
Fkbp11
Gpd1
Kansl2
Kcnh3
Kmt2d
Larp4
Letmd1
Lima1
Lmbr1l
LOC100362820
LOC102546778
LOC102548155
LOC102556302
LOC108351553
LOC108351554
LOC120093732
LOC120093735
LOC120093830
LOC120093832
Mcrs1
Mettl7a
Nckap5l
Olr1877
Pfkm
Prkag1
Prpf40b
Prph
Rhebl1
Rnd1
Senp1
Slc11a2
Slc48a1
Slc4a8
Smagp
Smarcd1
Spats2
Tmbim6
Tmprss12
Tuba1a
Tuba1b
Tuba1c
Wnt1
Zfp641
|
|
Tibialis anterior weight
|
4.56 |
3 |
0 |
0 |
1 |
4.95e-35 |
Letmd1
Mettl7a
Slc11a2
|
|
Tibia length
|
7.23 |
60 |
0 |
0 |
1 |
0.00e+00 |
Adcy6
Aqp5
Asb8
Asic1
Atf1
Bcdin3d
Bin2
C1ql4
Cacnb3
Ccdc65
Ccnt1
Cela1
Csrnp2
Ddn
Ddx23
Dhh
Dip2b
Dnajc22
Faim2
Fkbp11
Fmnl3
Kansl2
Kcnh3
Kmt2d
Lmbr1l
LOC100362820
LOC102546778
LOC102548155
LOC102555574
LOC102556302
LOC103692978
LOC108351553
LOC108351554
LOC120093732
LOC120093830
LOC120093831
Mcrs1
Mettl7a
Nckap5l
Olr1877
Pfkm
Prkag1
Prpf40b
Prph
Rhebl1
Rnd1
Senp1
Slc11a2
Slc48a1
Smagp
Smarcd1
Spats2
Tmbim6
Tmprss12
Tuba1a
Tuba1b
Tuba1c
Wnt1
Wnt10b
Zfp641
|
|
Bone: apparent density
|
10.68 |
49 |
0 |
0 |
1 |
0.00e+00 |
Adcy6
Asb8
Asic1
Bcdin3d
C1ql4
Cacnb3
Ccdc65
Ccnt1
Cela1
Ddn
Ddx23
Dhh
Dnajc22
Faim2
Fkbp11
Fmnl3
Kansl2
Kcnh3
Kmt2d
Lmbr1l
LOC100362820
LOC102546778
LOC102548155
LOC102555574
LOC102556302
LOC103692978
LOC108351554
LOC120093732
LOC120093830
LOC120093831
Mcrs1
Nckap5l
Olr1877
Pfkm
Prkag1
Prpf40b
Prph
Rhebl1
Rnd1
Senp1
Slc48a1
Spats2
Tmbim6
Tuba1a
Tuba1b
Tuba1c
Wnt1
Wnt10b
Zfp641
|
|
Bone surface
|
11.4 |
68 |
0 |
0 |
1 |
0.00e+00 |
Adcy6
Aqp5
Asb8
Asic1
Atf1
Bcdin3d
Bin2
C1ql4
Cacnb3
Ccdc65
Ccnt1
Cela1
Cers5
Csrnp2
Ddn
Ddx23
Dhh
Dip2b
Dnajc22
Faim2
Fkbp11
Fmnl3
Gpd1
Kansl2
Kcnh3
Kmt2d
Larp4
Letmd1
Lima1
Lmbr1l
LOC100362820
LOC102546778
LOC102548155
LOC102555574
LOC102556302
LOC103692978
LOC108351553
LOC108351554
LOC120093732
LOC120093735
LOC120093830
LOC120093831
LOC120093832
Mcrs1
Mettl7a
Nckap5l
Olr1877
Pfkm
Prkag1
Prpf40b
Prph
Rhebl1
Rnd1
Senp1
Slc11a2
Slc48a1
Slc4a8
Smagp
Smarcd1
Spats2
Tmbim6
Tmprss12
Tuba1a
Tuba1b
Tuba1c
Wnt1
Wnt10b
Zfp641
|
|
Bone volume
|
9.61 |
65 |
0 |
0 |
1 |
0.00e+00 |
Adcy6
Aqp5
Asic1
Atf1
Bcdin3d
Bin2
Cacnb3
Ccdc65
Ccnt1
Cela1
Cers5
Csrnp2
Ddn
Ddx23
Dhh
Dip2b
Dnajc22
Faim2
Fkbp11
Fmnl3
Gpd1
Kansl2
Kcnh3
Kmt2d
Larp4
Letmd1
Lima1
Lmbr1l
LOC100362820
LOC102546778
LOC102548155
LOC102555574
LOC102556302
LOC103692978
LOC108351553
LOC108351554
LOC120093732
LOC120093735
LOC120093830
LOC120093831
LOC120093832
Mcrs1
Mettl7a
Nckap5l
Olr1877
Pfkm
Prkag1
Prpf40b
Prph
Rhebl1
Rnd1
Senp1
Slc11a2
Slc48a1
Smagp
Smarcd1
Spats2
Tmbim6
Tmprss12
Tuba1a
Tuba1b
Tuba1c
Wnt1
Wnt10b
Zfp641
|
|
Bone: connectivity density
|
9.99 |
2 |
0 |
0 |
0 |
1.00e+00 |
Tuba1a
Tuba1b
|
|
Bone: cortical area
|
10.61 |
29 |
0 |
0 |
1 |
7.88e-182 |
Adcy6
Asb8
Cacnb3
Ccdc65
Ccnt1
Ddx23
Dhh
Fkbp11
Kansl2
Kcnh3
Kmt2d
Lmbr1l
LOC100362820
LOC102548155
LOC102556302
LOC120093732
LOC120093830
Olr1877
Pfkm
Prkag1
Rhebl1
Rnd1
Senp1
Slc48a1
Tuba1a
Tuba1b
Tuba1c
Wnt1
Zfp641
|
|
Bone: endosteal estimation
|
7.9 |
68 |
0 |
0 |
1 |
0.00e+00 |
Adcy6
Aqp5
Asb8
Asic1
Atf1
Bcdin3d
Bin2
C1ql4
Cacnb3
Ccdc65
Ccnt1
Cela1
Cers5
Csrnp2
Ddn
Ddx23
Dhh
Dip2b
Dnajc22
Faim2
Fkbp11
Fmnl3
Gpd1
Kansl2
Kcnh3
Kmt2d
Larp4
Letmd1
Lima1
Lmbr1l
LOC100362820
LOC102546778
LOC102548155
LOC102555574
LOC102556302
LOC103692978
LOC108351553
LOC108351554
LOC120093732
LOC120093735
LOC120093830
LOC120093831
LOC120093832
Mcrs1
Mettl7a
Nckap5l
Olr1877
Pfkm
Prkag1
Prpf40b
Prph
Rhebl1
Rnd1
Senp1
Slc11a2
Slc48a1
Slc4a8
Smagp
Smarcd1
Spats2
Tmbim6
Tmprss12
Tuba1a
Tuba1b
Tuba1c
Wnt1
Wnt10b
Zfp641
|
|
Bone: endosteal perimeter
|
7.58 |
40 |
0 |
0 |
1 |
1.34e-152 |
Adcy6
Aqp5
Asb8
Asic1
Atf1
Bcdin3d
Bin2
Cacnb3
Cela1
Csrnp2
Dip2b
Dnajc22
Faim2
Kansl2
Kcnh3
Larp4
Letmd1
Lmbr1l
LOC100362820
LOC102546778
LOC108351553
LOC108351554
LOC120093830
Mettl7a
Nckap5l
Olr1877
Pfkm
Prkag1
Rnd1
Slc11a2
Slc48a1
Smagp
Smarcd1
Spats2
Tmbim6
Tmprss12
Tuba1a
Tuba1b
Tuba1c
Zfp641
|
|
Bone: final force
|
10.77 |
68 |
0 |
0 |
1 |
0.00e+00 |
Adcy6
Aqp5
Asic1
Atf1
Bcdin3d
Bin2
C1ql4
Cacnb3
Ccdc65
Ccnt1
Cela1
Cers5
Cox14
Csrnp2
Ddn
Ddx23
Dhh
Dip2b
Dnajc22
Faim2
Fkbp11
Fmnl3
Gpd1
Kansl2
Kcnh3
Kmt2d
Larp4
Letmd1
Lima1
Lmbr1l
LOC100362820
LOC102546778
LOC102548155
LOC102555574
LOC102556302
LOC103692978
LOC108351553
LOC108351554
LOC120093732
LOC120093735
LOC120093830
LOC120093831
LOC120093832
Mcrs1
Mettl7a
Nckap5l
Olr1877
Pfkm
Prkag1
Prpf40b
Prph
Rhebl1
Rnd1
Senp1
Slc11a2
Slc48a1
Slc4a8
Smagp
Smarcd1
Spats2
Tmbim6
Tmprss12
Tuba1a
Tuba1b
Tuba1c
Wnt1
Wnt10b
Zfp641
|
|
Bone: final moment
|
11.73 |
69 |
0 |
0 |
1 |
0.00e+00 |
Adcy6
Aqp5
Asb8
Asic1
Atf1
Bcdin3d
Bin2
C1ql4
Cacnb3
Ccdc65
Ccnt1
Cela1
Cers5
Cox14
Csrnp2
Ddn
Ddx23
Dhh
Dip2b
Dnajc22
Faim2
Fkbp11
Fmnl3
Gpd1
Kansl2
Kcnh3
Kmt2d
Larp4
Letmd1
Lima1
Lmbr1l
LOC100362820
LOC102546778
LOC102548155
LOC102555574
LOC102556302
LOC103692978
LOC108351553
LOC108351554
LOC120093732
LOC120093735
LOC120093830
LOC120093831
LOC120093832
Mcrs1
Mettl7a
Nckap5l
Olr1877
Pfkm
Prkag1
Prpf40b
Prph
Rhebl1
Rnd1
Senp1
Slc11a2
Slc48a1
Slc4a8
Smagp
Smarcd1
Spats2
Tmbim6
Tmprss12
Tuba1a
Tuba1b
Tuba1c
Wnt1
Wnt10b
Zfp641
|
|
Bone: marrow area
|
7.8 |
62 |
0 |
0 |
1 |
0.00e+00 |
Adcy6
Aqp5
Asb8
Asic1
Atf1
Bcdin3d
Bin2
C1ql4
Cacnb3
Ccdc65
Ccnt1
Cela1
Cers5
Csrnp2
Ddn
Ddx23
Dhh
Dip2b
Dnajc22
Faim2
Fkbp11
Fmnl3
Kansl2
Kcnh3
Kmt2d
Larp4
Letmd1
Lima1
Lmbr1l
LOC100362820
LOC102546778
LOC102555574
LOC102556302
LOC103692978
LOC108351553
LOC108351554
LOC120093830
LOC120093831
Mcrs1
Mettl7a
Nckap5l
Olr1877
Pfkm
Prkag1
Prpf40b
Prph
Rhebl1
Rnd1
Slc11a2
Slc48a1
Slc4a8
Smagp
Smarcd1
Spats2
Tmbim6
Tmprss12
Tuba1a
Tuba1b
Tuba1c
Wnt1
Wnt10b
Zfp641
|
|
Bone: maximum force
|
14.57 |
69 |
22 |
314.3 |
1 |
0.00e+00 |
Adcy6
Aqp5
Asb8
Asic1
Atf1
Bcdin3d
Bin2
C1ql4
Cacnb3
Ccdc65
Ccnt1
Cela1
Cers5
Cox14
Csrnp2
Ddn
Ddx23
Dhh
Dip2b
Dnajc22
Faim2
Fkbp11
Fmnl3
Gpd1
Kansl2
Kcnh3
Kmt2d
Larp4
Letmd1
Lima1
Lmbr1l
LOC100362820
LOC102546778
LOC102548155
LOC102555574
LOC102556302
LOC103692978
LOC108351553
LOC108351554
LOC120093732
LOC120093735
LOC120093830
LOC120093831
LOC120093832
Mcrs1
Mettl7a
Nckap5l
Olr1877
Pfkm
Prkag1
Prpf40b
Prph
Rhebl1
Rnd1
Senp1
Slc11a2
Slc48a1
Slc4a8
Smagp
Smarcd1
Spats2
Tmbim6
Tmprss12
Tuba1a
Tuba1b
Tuba1c
Wnt1
Wnt10b
Zfp641
|
|
Bone: maximum moment
|
15.7 |
69 |
55 |
785.7 |
1 |
0.00e+00 |
Adcy6
Aqp5
Asb8
Asic1
Atf1
Bcdin3d
Bin2
C1ql4
Cacnb3
Ccdc65
Ccnt1
Cela1
Cers5
Cox14
Csrnp2
Ddn
Ddx23
Dhh
Dip2b
Dnajc22
Faim2
Fkbp11
Fmnl3
Gpd1
Kansl2
Kcnh3
Kmt2d
Larp4
Letmd1
Lima1
Lmbr1l
LOC100362820
LOC102546778
LOC102548155
LOC102555574
LOC102556302
LOC103692978
LOC108351553
LOC108351554
LOC120093732
LOC120093735
LOC120093830
LOC120093831
LOC120093832
Mcrs1
Mettl7a
Nckap5l
Olr1877
Pfkm
Prkag1
Prpf40b
Prph
Rhebl1
Rnd1
Senp1
Slc11a2
Slc48a1
Slc4a8
Smagp
Smarcd1
Spats2
Tmbim6
Tmprss12
Tuba1a
Tuba1b
Tuba1c
Wnt1
Wnt10b
Zfp641
|
|
Bone: minimum diameter
|
9.07 |
66 |
0 |
0 |
1 |
0.00e+00 |
Adcy6
Aqp5
Asb8
Asic1
Atf1
Bcdin3d
Bin2
C1ql4
Cacnb3
Ccdc65
Ccnt1
Cers5
Csrnp2
Ddn
Ddx23
Dhh
Dip2b
Dnajc22
Faim2
Fkbp11
Fmnl3
Gpd1
Kansl2
Kcnh3
Kmt2d
Larp4
Letmd1
Lima1
Lmbr1l
LOC100362820
LOC102546778
LOC102548155
LOC102555574
LOC102556302
LOC103692978
LOC108351553
LOC108351554
LOC120093732
LOC120093735
LOC120093830
LOC120093831
LOC120093832
Mcrs1
Mettl7a
Nckap5l
Olr1877
Pfkm
Prkag1
Prpf40b
Prph
Rhebl1
Rnd1
Senp1
Slc11a2
Slc48a1
Smagp
Smarcd1
Spats2
Tmbim6
Tmprss12
Tuba1a
Tuba1b
Tuba1c
Wnt1
Wnt10b
Zfp641
|
|
Bone: periosteal estimation
|
11.03 |
69 |
12 |
171.4 |
1 |
0.00e+00 |
Adcy6
Aqp5
Asb8
Asic1
Atf1
Bcdin3d
Bin2
C1ql4
Cacnb3
Ccdc65
Ccnt1
Cela1
Cers5
Cox14
Csrnp2
Ddn
Ddx23
Dhh
Dip2b
Dnajc22
Faim2
Fkbp11
Fmnl3
Gpd1
Kansl2
Kcnh3
Kmt2d
Larp4
Letmd1
Lima1
Lmbr1l
LOC100362820
LOC102546778
LOC102548155
LOC102555574
LOC102556302
LOC103692978
LOC108351553
LOC108351554
LOC120093732
LOC120093735
LOC120093830
LOC120093831
LOC120093832
Mcrs1
Mettl7a
Nckap5l
Olr1877
Pfkm
Prkag1
Prpf40b
Prph
Rhebl1
Rnd1
Senp1
Slc11a2
Slc48a1
Slc4a8
Smagp
Smarcd1
Spats2
Tmbim6
Tmprss12
Tuba1a
Tuba1b
Tuba1c
Wnt1
Wnt10b
Zfp641
|
|
Bone: periosteal perimeter
|
10.21 |
67 |
0 |
0 |
1 |
0.00e+00 |
Adcy6
Aqp5
Asb8
Asic1
Atf1
Bcdin3d
Bin2
C1ql4
Cacnb3
Ccdc65
Ccnt1
Cela1
Cers5
Csrnp2
Ddn
Ddx23
Dhh
Dip2b
Dnajc22
Faim2
Fkbp11
Fmnl3
Kansl2
Kcnh3
Kmt2d
Larp4
Letmd1
Lima1
Lmbr1l
LOC100362820
LOC102546778
LOC102548155
LOC102555574
LOC102556302
LOC103692978
LOC108351553
LOC108351554
LOC120093732
LOC120093735
LOC120093830
LOC120093831
LOC120093832
Mcrs1
Mettl7a
Nckap5l
Olr1877
Pfkm
Prkag1
Prpf40b
Prph
Rhebl1
Rnd1
Senp1
Slc11a2
Slc48a1
Slc4a8
Smagp
Smarcd1
Spats2
Tmbim6
Tmprss12
Tuba1a
Tuba1b
Tuba1c
Wnt1
Wnt10b
Zfp641
|
|
Bone: post-yield work
|
11.67 |
2 |
0 |
0 |
0 |
1.00e+00 |
Dip2b
LOC100362820
|
|
Bone: trabecular number
|
9.79 |
2 |
0 |
0 |
0 |
1.00e+00 |
Tuba1a
Tuba1b
|