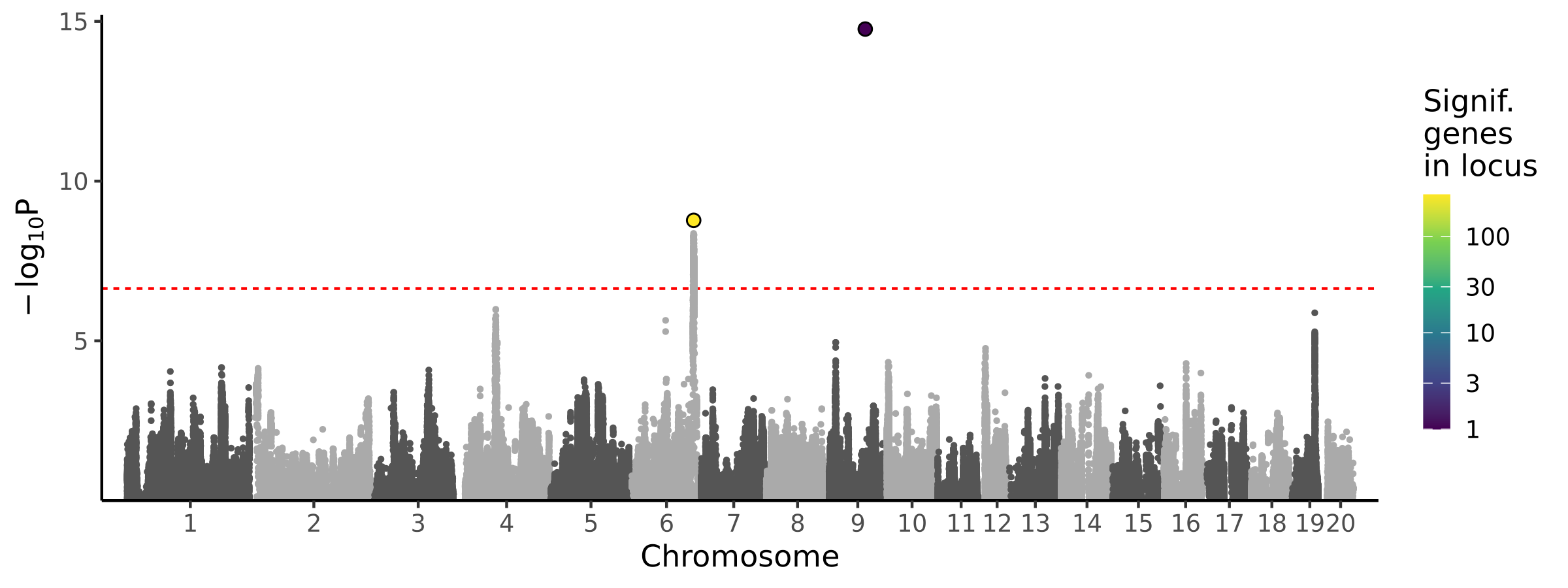
|
BMI with tail
|
8.42 |
1 |
1 |
8.3 |
0 |
1.00e+00 |
Zfand2b
|
|
BMI without tail
|
53.65 |
1 |
1 |
8.3 |
0 |
1.00e+00 |
Zfand2b
|
|
Epididymis fat weight
|
111.28 |
1 |
1 |
8.3 |
0 |
1.00e+00 |
Zfand2b
|
|
Heart weight
|
71.7 |
1 |
1 |
8.3 |
0 |
1.00e+00 |
Zfand2b
|
|
Left kidney weight
|
27.22 |
1 |
1 |
8.3 |
0 |
1.00e+00 |
Zfand2b
|
|
Right kidney weight
|
54.05 |
1 |
1 |
8.3 |
0 |
1.00e+00 |
Zfand2b
|
|
Tail length
|
6.08 |
23 |
1 |
8.3 |
0.89 |
4.62e-62 |
Ahnak2
Brf1
Btbd6
C6h14orf180
Cdca4
Cep170b
Clba1
Crip1
Crip2
Gpr132
Inf2
Jag2
LOC108351305
LOC120103631
LOC120103632
Nudt14
Pacs2
Pld4
Siva1
Tedc1
Tex22
Tmem121
Zfand2b
|
|
Length with tail
|
7.6 |
47 |
22 |
183.3 |
0.98 |
3.88e-289 |
Ahnak2
Amn
Aspg
Bag5
Brf1
Btbd6
C6h14orf180
Cdc42bpb
Cdca4
Cep170b
Ckb
Clba1
Coa8
Crip1
Crip2
Diras3
Eif5
Exoc3l4
Gpr132
Inf2
Jag2
Kif26a
Klc1
Lbhd2
LOC108351305
LOC120103628
LOC120103629
LOC120103631
LOC120103632
Mark3
Nudt14
Pacs2
Pld4
Ppp1r13b
Rd3l
Siva1
Snora28l1
Tdrd9
Tedc1
Tex22
Tmem121
Tmem179
Tnfaip2
Traf3
Xrcc3
Zfand2b
Zfyve21
|
|
Length without tail
|
11.93 |
8 |
1 |
8.3 |
0.95 |
9.33e-08 |
Ahnak2
Brf1
Cep170b
Jag2
Nudt14
Pacs2
Tedc1
Zfand2b
|
|
Liver weight, right
|
17.57 |
1 |
1 |
8.3 |
0 |
1.00e+00 |
Zfand2b
|
|
Parametrial fat weight
|
9.99 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zfand2b
|
|
Retroperitoneal fat weight
|
7.53 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zfand2b
|
|
Intraocular pressure
|
51.69 |
1 |
1 |
8.3 |
0 |
1.00e+00 |
Zfand2b
|
|
Extensor digitorum longus weight
|
7.81 |
49 |
31 |
258.3 |
0.99 |
0.00e+00 |
Ahnak2
Amn
Aspg
Bag5
Brf1
Btbd6
C6h14orf180
Cdc42bpb
Cdca4
Cep170b
Cinp
Ckb
Clba1
Coa8
Crip1
Crip2
Diras3
Eif5
Exoc3l4
Gpr132
Inf2
Jag2
Kif26a
Klc1
Lbhd2
LOC108351305
LOC120103628
LOC120103629
LOC120103631
LOC120103632
Mark3
Nudt14
Pacs2
Pld4
Ppp1r13b
Rd3l
Siva1
Snora28l1
Tdrd9
Tedc1
Tex22
Tmem121
Tmem179
Tnfaip2
Traf3
Trmt61a
Wdr20
Xrcc3
Zfyve21
|
|
Number of licking bursts
|
32.5 |
1 |
1 |
8.3 |
0 |
1.00e+00 |
Zfand2b
|
|
Food consumed during 24 hour testing period
|
15.78 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zfand2b
|
|
Mean num. licks in bursts
|
18.13 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zfand2b
|
|
Water consumed over 24 hours
|
34.33 |
1 |
1 |
8.3 |
0 |
1.00e+00 |
Zfand2b
|
|
Patch foraging indifference point 0 sec
|
11.8 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zfand2b
|
|
Patch foraging indifference point AUC
|
24.2 |
1 |
1 |
8.3 |
0 |
1.00e+00 |
Zfand2b
|
|
Indifference point function ln k
|
54.42 |
1 |
1 |
8.3 |
0 |
1.00e+00 |
Zfand2b
|
|
Indifference point function log k
|
54.42 |
1 |
1 |
8.3 |
0 |
1.00e+00 |
Zfand2b
|
|
Patch foraging total patch changes 0 sec
|
10.98 |
8 |
1 |
8.3 |
0.7 |
8.15e-03 |
Brf1
Cep170b
Crip2
Jag2
Nudt14
Tedc1
Tmem121
Zfand2b
|
|
Patch foraging total patch changes 18 sec
|
48.28 |
1 |
1 |
8.3 |
0 |
1.00e+00 |
Zfand2b
|
|
Patch foraging total patch changes 24 sec
|
77.61 |
1 |
1 |
8.3 |
0 |
1.00e+00 |
Zfand2b
|
|
Patch foraging total patch changes 6 sec
|
13.35 |
5 |
1 |
8.3 |
0.45 |
3.67e-01 |
Crip2
Jag2
Tedc1
Tmem121
Zfand2b
|
|
Patch foraging time to switch 0 sec
|
11.92 |
49 |
1 |
8.3 |
-1 |
0.00e+00 |
Ahnak2
Amn
Aspg
Bag5
Brf1
Btbd6
C6h14orf180
Cdc42bpb
Cdca4
Cep170b
Cinp
Ckb
Clba1
Coa8
Crip1
Crip2
Diras3
Eif5
Exoc3l4
Gpr132
Inf2
Jag2
Kif26a
Klc1
Lbhd2
LOC108351305
LOC120103628
LOC120103629
LOC120103631
LOC120103632
Mark3
Nudt14
Pacs2
Pld4
Ppp1r13b
Rd3l
Siva1
Snora28l1
Tdrd9
Tedc1
Tex22
Tmem121
Tmem179
Tnfaip2
Traf3
Trmt61a
Xrcc3
Zfand2b
Zfyve21
|
|
Patch foraging water rate 12 sec
|
11.7 |
26 |
1 |
8.3 |
0.99 |
1.04e-89 |
Aspg
Bag5
Brf1
C6h14orf180
Cdc42bpb
Cinp
Coa8
Eif5
Kif26a
Klc1
LOC108351305
LOC120103628
LOC120103631
LOC120103632
Ppp1r13b
Rd3l
Siva1
Tdrd9
Tmem179
Tnfaip2
Traf3
Trmt61a
Wdr20
Xrcc3
Zfand2b
Zfyve21
|
|
Patch foraging water rate 24 sec
|
12.61 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zfand2b
|
|
Patch foraging water rate 6 sec
|
35.43 |
1 |
1 |
8.3 |
0 |
1.00e+00 |
Zfand2b
|
|
Locomotor activity
|
12.38 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zfand2b
|
|
Locomotor testing distance
|
47.2 |
1 |
1 |
8.3 |
0 |
1.00e+00 |
Zfand2b
|
|
Locomotor testing rearing
|
99.51 |
1 |
1 |
8.3 |
0 |
1.00e+00 |
Zfand2b
|
|
Light reinforcement 1
|
19.25 |
1 |
1 |
8.3 |
0 |
1.00e+00 |
Zfand2b
|
|
Reaction time number correct
|
114.79 |
1 |
1 |
8.3 |
0 |
1.00e+00 |
Zfand2b
|
|
Reaction time mean minus median
|
179.03 |
1 |
1 |
8.3 |
0 |
1.00e+00 |
Zfand2b
|
|
Reaction time mean minus median AUC
|
232.47 |
1 |
1 |
8.3 |
0 |
1.00e+00 |
Zfand2b
|
|
Reaction time num false alarms AUC
|
27.43 |
1 |
1 |
8.3 |
0 |
1.00e+00 |
Zfand2b
|
|
Reaction time trials correct on left
|
114.79 |
1 |
1 |
8.3 |
0 |
1.00e+00 |
Zfand2b
|
|
Reaction time trials on left
|
106.69 |
1 |
1 |
8.3 |
0 |
1.00e+00 |
Zfand2b
|
|
Reaction time mean
|
36.11 |
1 |
1 |
8.3 |
0 |
1.00e+00 |
Zfand2b
|
|
Reaction time mean AUC
|
33.75 |
1 |
1 |
8.3 |
0 |
1.00e+00 |
Zfand2b
|
|
Median of all reaction times
|
17.65 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zfand2b
|
|
Reaction time omissions
|
46.56 |
1 |
1 |
8.3 |
0 |
1.00e+00 |
Zfand2b
|
|
Reaction time premature initiations
|
41.6 |
1 |
1 |
8.3 |
0 |
1.00e+00 |
Zfand2b
|
|
Std. dev. reaction times
|
72.92 |
1 |
1 |
8.3 |
0 |
1.00e+00 |
Zfand2b
|
|
Reaction time trials completed
|
106.69 |
1 |
1 |
8.3 |
0 |
1.00e+00 |
Zfand2b
|
|
Reaction time trials AUC
|
108.76 |
1 |
1 |
8.3 |
0 |
1.00e+00 |
Zfand2b
|
|
Conditioned reinforcement - actives
|
152.54 |
1 |
1 |
8.3 |
0 |
1.00e+00 |
Zfand2b
|
|
Conditioned reinforcement - inactives
|
135.63 |
1 |
1 |
8.3 |
0 |
1.00e+00 |
Zfand2b
|
|
Conditioned locomotion
|
13.93 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zfand2b
|
|
Intermittent access intake day 1-15 change
|
156.77 |
1 |
1 |
8.3 |
0 |
1.00e+00 |
Zfand2b
|
|
Intermittent access intake escalation
|
117.41 |
1 |
1 |
8.3 |
0 |
1.00e+00 |
Zfand2b
|
|
Intermitt. access day 1 inactive lever presses
|
169.63 |
1 |
1 |
8.3 |
0 |
1.00e+00 |
Zfand2b
|
|
Intermitt. access day 15 inactive lever presses
|
294.26 |
1 |
1 |
8.3 |
0 |
1.00e+00 |
Zfand2b
|
|
Intermittent access day 1 total infusions
|
71.42 |
1 |
1 |
8.3 |
0 |
1.00e+00 |
Zfand2b
|
|
Intermittent access day 15 total infusions
|
35.39 |
1 |
1 |
8.3 |
0 |
1.00e+00 |
Zfand2b
|
|
Intermittent access terminal intake (last 3 days)
|
25.82 |
1 |
1 |
8.3 |
0 |
1.00e+00 |
Zfand2b
|
|
Intermittent access day 1 locomotion
|
26.47 |
1 |
1 |
8.3 |
0 |
1.00e+00 |
Zfand2b
|
|
Intermittent access day 15 total locomotion
|
93.08 |
1 |
1 |
8.3 |
0 |
1.00e+00 |
Zfand2b
|
|
Intermittent access standard deviation
|
442.51 |
1 |
1 |
8.3 |
0 |
1.00e+00 |
Zfand2b
|
|
Cocaine induced anxiety
|
125.35 |
1 |
1 |
8.3 |
0 |
1.00e+00 |
Zfand2b
|
|
Baseline Anxiety
|
393.92 |
1 |
1 |
8.3 |
0 |
1.00e+00 |
Zfand2b
|
|
Incentive Sensitization - Responses
|
309.83 |
1 |
1 |
8.3 |
0 |
1.00e+00 |
Zfand2b
|
|
Incentive Sensitization - Breakpoint
|
300.45 |
1 |
1 |
8.3 |
0 |
1.00e+00 |
Zfand2b
|
|
Progressive ratio test 1 active lever presses
|
26.41 |
1 |
1 |
8.3 |
0 |
1.00e+00 |
Zfand2b
|
|
Progressive ratio test 1 breakpoint
|
19.19 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zfand2b
|
|
Progressive ratio test 2 active lever presses
|
109.22 |
1 |
1 |
8.3 |
0 |
1.00e+00 |
Zfand2b
|
|
Progressive ratio test 2 breakpoint
|
106.65 |
1 |
1 |
8.3 |
0 |
1.00e+00 |
Zfand2b
|
|
Progressive ratio test 2 inactive lever presses
|
63.47 |
1 |
1 |
8.3 |
0 |
1.00e+00 |
Zfand2b
|
|
Short access day 1 total inactive lever presses
|
21.93 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zfand2b
|
|
Short access day 10 total inactive lever presses
|
43.04 |
1 |
1 |
8.3 |
0 |
1.00e+00 |
Zfand2b
|
|
Short access day 1 locomotion
|
129.19 |
1 |
1 |
8.3 |
0 |
1.00e+00 |
Zfand2b
|
|
Short access day 10 total locomotion
|
432.47 |
1 |
1 |
8.3 |
0 |
1.00e+00 |
Zfand2b
|
|
Short access total locomotion
|
209.96 |
1 |
1 |
8.3 |
0 |
1.00e+00 |
Zfand2b
|
|
Compulsive drug intake
|
68.39 |
1 |
1 |
8.3 |
0 |
1.00e+00 |
Zfand2b
|
|
One hour access (0.3 mA shock)
|
22.58 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zfand2b
|
|
Number of responses in last shaping day
|
67.75 |
1 |
1 |
8.3 |
0 |
1.00e+00 |
Zfand2b
|
|
Delta time in open arm before/after self-admin
|
26.67 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zfand2b
|
|
Active lever presses in extinction session 6
|
26.41 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zfand2b
|
|
Time in closed arm before self-admin
|
44.32 |
1 |
1 |
8.3 |
0 |
1.00e+00 |
Zfand2b
|
|
Time in open arm before self-admin
|
24.67 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zfand2b
|
|
Time to tail flick, vehicle, after self-admin
|
75.62 |
1 |
1 |
8.3 |
0 |
1.00e+00 |
Zfand2b
|
|
Delta time to tail flick, vehicle, before/after SA
|
74.6 |
1 |
1 |
8.3 |
0 |
1.00e+00 |
Zfand2b
|
|
Ambulatory time before self-admin
|
25.36 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zfand2b
|
|
Area under the delay curve
|
31.15 |
1 |
1 |
8.3 |
0 |
1.00e+00 |
Zfand2b
|
|
Delay disc. indifference point, 16s delay
|
69.03 |
1 |
1 |
8.3 |
0 |
1.00e+00 |
Zfand2b
|
|
Delay disc. indifference point, 24s delay
|
27.43 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zfand2b
|
|
Delay disc. indifference point, 8s delay
|
74.24 |
1 |
1 |
8.3 |
0 |
1.00e+00 |
Zfand2b
|
|
Delay discount exponential model param
|
16.14 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zfand2b
|
|
Delay discount hyperbolic model param
|
15.89 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zfand2b
|
|
Fecal boli incidents, locomotor time 1
|
176.47 |
1 |
1 |
8.3 |
0 |
1.00e+00 |
Zfand2b
|
|
Time >=10cm from walls, locomotor time 1
|
79.04 |
1 |
1 |
8.3 |
0 |
1.00e+00 |
Zfand2b
|
|
Vertical activity count, locomotor time 1
|
97.7 |
1 |
1 |
8.3 |
0 |
1.00e+00 |
Zfand2b
|
|
Bouts of movement, locomotor time 2
|
23.9 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zfand2b
|
|
Total resting periods, locomotor time 2
|
36.28 |
1 |
1 |
8.3 |
0 |
1.00e+00 |
Zfand2b
|
|
Rest time, locomotor task time 2
|
44.91 |
1 |
1 |
8.3 |
0 |
1.00e+00 |
Zfand2b
|
|
Distance moved, locomotor task time 2
|
83.87 |
1 |
1 |
8.3 |
0 |
1.00e+00 |
Zfand2b
|
|
Locomotion in novel chamber post-restriction
|
14.04 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zfand2b
|
|
Food seeking constrained by brief footshock
|
14.98 |
1 |
0 |
0 |
0 |
1.00e+00 |
Zfand2b
|
|
Run reversals in cocaine runway, females
|
36.45 |
1 |
1 |
8.3 |
0 |
1.00e+00 |
Zfand2b
|
|
Latency to leave start box in cocaine runway
|
34.23 |
1 |
1 |
8.3 |
0 |
1.00e+00 |
Zfand2b
|
|
Latency to leave start box in cocaine runway, F
|
31.9 |
1 |
1 |
8.3 |
0 |
1.00e+00 |
Zfand2b
|