Delay disc. indifference point, 24s delay
Abnormal impulse behavior control/Impulsivity assay, delay discounting using an adjusting amount procedure: subject's calculated indifference point for all exposures/repetitions of 24s delay sessions in 11-16 week old NIH heterogeneous stock rats for males and females (MEDPC V equipment, where immediate lever with 0s delay provides choice dependent adjusting amounts starting at 75 microliter of sucrose solution and delayed lever provides 150 microliter of sucrose solution) [microliter].
Tags: Behavior · Impulsivity · Delay discounting
Project: u01_suzanne_mitchell
0 loci · 0 genes with independent associations · 0 total associated genes
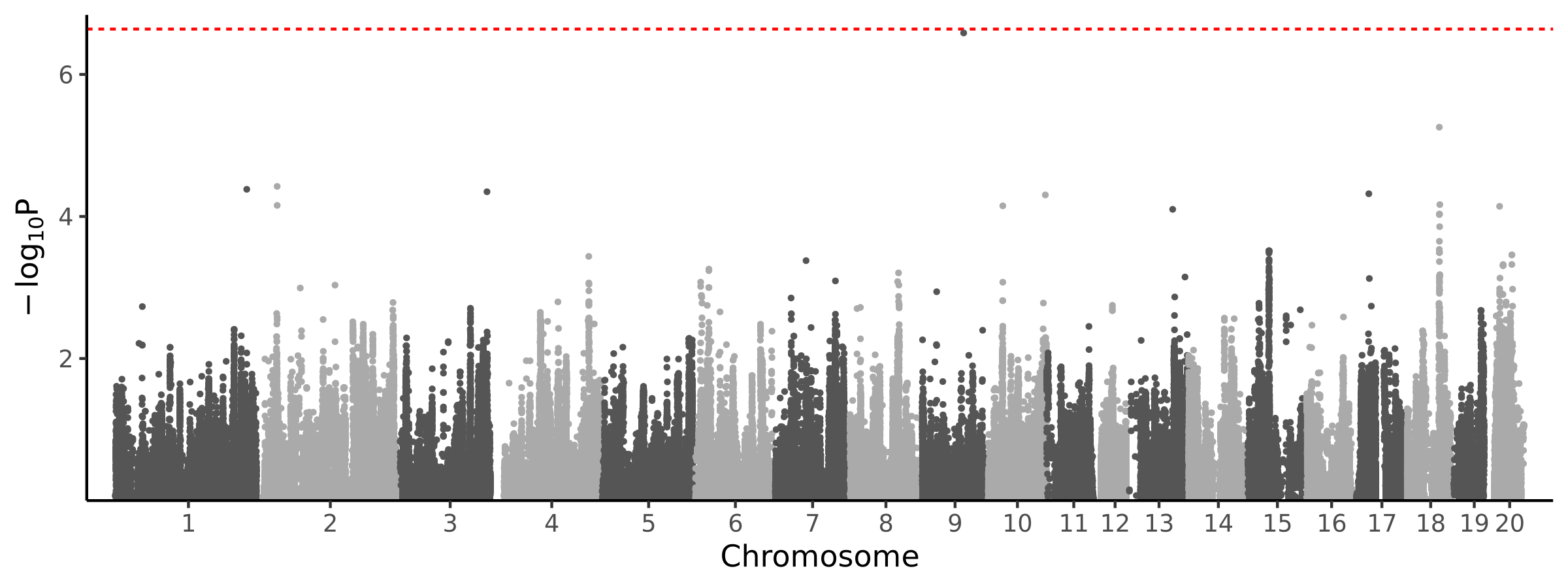
Significant Loci
| # | Chr | Start pos | End pos | # assoc genes | # joint models | Best TWAS P | Best GWAS P | Cond GWAS P | Joint genes |
|---|
Pleiotropic Associations
| Trait | Chi2 ratio | # genes+ | # genes++ | % genes++ | Corr | Corr P | Genes |
|---|
Associations by panel
| Tissue | RNA modality | # hits | % hits/tests | Avg chisq |
|---|---|---|---|---|
| Adipose | alternative polyA | 0 | 0 | 0.96 |
| Adipose | alternative TSS | 0 | 0 | 0.87 |
| Adipose | gene expression | 0 | 0 | 0.99 |
| Adipose | isoform ratio | 0 | 0 | 0.95 |
| Adipose | intron excision ratio | 0 | 0 | 0.88 |
| Adipose | mRNA stability | 0 | 0 | 0.98 |
| BLA | alternative polyA | 0 | 0 | 0.95 |
| BLA | alternative TSS | 0 | 0 | 0.92 |
| BLA | gene expression | 0 | 0 | 0.98 |
| BLA | isoform ratio | 0 | 0 | 0.95 |
| BLA | intron excision ratio | 0 | 0 | 1 |
| BLA | mRNA stability | 0 | 0 | 1 |
| Brain | alternative polyA | 0 | 0 | 0.95 |
| Brain | alternative TSS | 0 | 0 | 0.95 |
| Brain | gene expression | 0 | 0 | 0.98 |
| Brain | isoform ratio | 0 | 0 | 0.95 |
| Brain | intron excision ratio | 0 | 0 | 0.95 |
| Brain | mRNA stability | 0 | 0 | 0.99 |
| Eye | alternative polyA | 0 | 0 | 0.95 |
| Eye | alternative TSS | 0 | 0 | 0.97 |
| Eye | gene expression | 0 | 0 | 0.98 |
| Eye | isoform ratio | 0 | 0 | 0.97 |
| Eye | intron excision ratio | 0 | 0 | 0.96 |
| Eye | mRNA stability | 0 | 0 | 0.9 |
| IL | alternative polyA | 0 | 0 | 0.98 |
| IL | alternative TSS | 0 | 0 | 0.91 |
| IL | gene expression | 0 | 0 | 0.97 |
| IL | isoform ratio | 0 | 0 | 0.89 |
| IL | intron excision ratio | 0 | 0 | 0.93 |
| IL | mRNA stability | 0 | 0 | 0.97 |
| LHb | alternative polyA | 0 | 0 | 1.02 |
| LHb | alternative TSS | 0 | 0 | 0.97 |
| LHb | gene expression | 0 | 0 | 0.95 |
| LHb | isoform ratio | 0 | 0 | 0.96 |
| LHb | intron excision ratio | 0 | 0 | 0.98 |
| LHb | mRNA stability | 0 | 0 | 0.95 |
| Liver | alternative polyA | 0 | 0 | 1.01 |
| Liver | alternative TSS | 0 | 0 | 0.99 |
| Liver | gene expression | 0 | 0 | 1.01 |
| Liver | isoform ratio | 0 | 0 | 0.98 |
| Liver | intron excision ratio | 0 | 0 | 1.08 |
| Liver | mRNA stability | 0 | 0 | 1.01 |
| NAcc | alternative polyA | 0 | 0 | 0.98 |
| NAcc | alternative TSS | 0 | 0 | 0.93 |
| NAcc | gene expression | 0 | 0 | 0.99 |
| NAcc | isoform ratio | 0 | 0 | 0.94 |
| NAcc | intron excision ratio | 0 | 0 | 0.95 |
| NAcc | mRNA stability | 0 | 0 | 0.98 |
| OFC | alternative polyA | 0 | 0 | 0.88 |
| OFC | alternative TSS | 0 | 0 | 0.89 |
| OFC | gene expression | 0 | 0 | 0.99 |
| OFC | isoform ratio | 0 | 0 | 0.94 |
| OFC | intron excision ratio | 0 | 0 | 0.89 |
| OFC | mRNA stability | 0 | 0 | 0.94 |
| PL | alternative polyA | 0 | 0 | 0.95 |
| PL | alternative TSS | 0 | 0 | 0.95 |
| PL | gene expression | 0 | 0 | 0.98 |
| PL | isoform ratio | 0 | 0 | 0.95 |
| PL | intron excision ratio | 0 | 0 | 0.94 |
| PL | mRNA stability | 0 | 0 | 0.98 |
| pVTA | alternative polyA | 0 | 0 | 1.03 |
| pVTA | alternative TSS | 0 | 0 | 0.96 |
| pVTA | gene expression | 0 | 0 | 1 |
| pVTA | isoform ratio | 0 | 0 | 0.96 |
| pVTA | intron excision ratio | 0 | 0 | 0.93 |
| pVTA | mRNA stability | 0 | 0 | 0.99 |
| RMTg | alternative polyA | 0 | 0 | 0.88 |
| RMTg | alternative TSS | 0 | 0 | 1.11 |
| RMTg | gene expression | 0 | 0 | 0.97 |
| RMTg | isoform ratio | 0 | 0 | 0.98 |
| RMTg | intron excision ratio | 0 | 0 | 0.93 |
| RMTg | mRNA stability | 0 | 0 | 0.95 |