Progressive ratio
Progressive ratio, i.e. food-seeking constrained by escalating effort schedule. The last successfully completed ratio or shock intensity endured before timeout. Animals are food restricted to 85% of initial body weight, trained to lever-press for food pellets (45 mg) until stable response rates were reached, then tested either on the progressive ratio (PR) task with progressively increasing effort requirements (lever-presses required per pellet) or progressively increasing punishment (i.e., a foot-shock of increasing intensity presented 1 s after food pellet delivery, whose magnitude increases ~25% every 3 trials). In the PR task, rats time out if 2 min passed without a lever press. In the punishment task, a timeout is defined by three consecutive trials with >30 s passing without a lever press. For both tasks, the break point is the last successfully completed ratio or shock intensity endured before timeout.
Tags: Behavior · Motivation
Project: u01_tom_jhou
0 loci · 0 genes with independent associations · 0 total associated genes
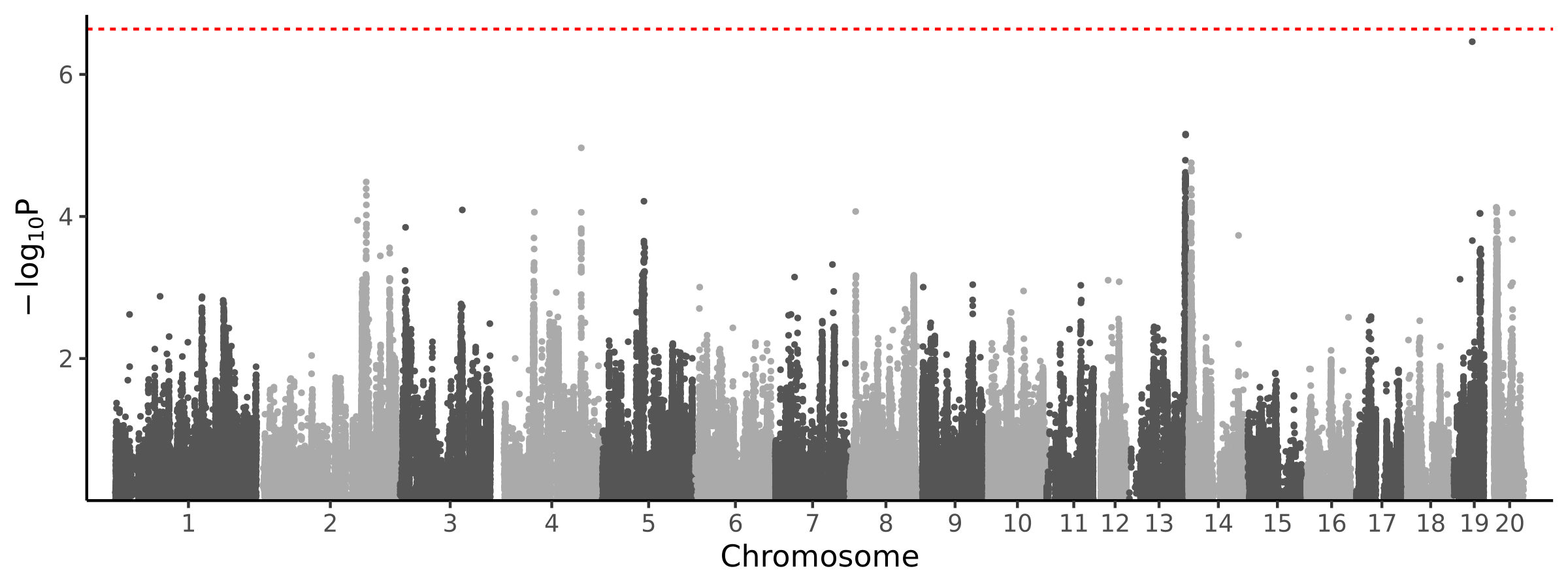
Significant Loci
| # | Chr | Start pos | End pos | # assoc genes | # joint models | Best TWAS P | Best GWAS P | Cond GWAS P | Joint genes |
|---|
Pleiotropic Associations
| Trait | Chi2 ratio | # genes+ | # genes++ | % genes++ | Corr | Corr P | Genes |
|---|
Associations by panel
| Tissue | RNA modality | # hits | % hits/tests | Avg chisq |
|---|---|---|---|---|
| Adipose | alternative polyA | 0 | 0 | 1.15 |
| Adipose | alternative TSS | 0 | 0 | 1.08 |
| Adipose | gene expression | 0 | 0 | 1.1 |
| Adipose | isoform ratio | 0 | 0 | 1.19 |
| Adipose | intron excision ratio | 0 | 0 | 1.21 |
| Adipose | mRNA stability | 0 | 0 | 1.14 |
| BLA | alternative polyA | 0 | 0 | 1.18 |
| BLA | alternative TSS | 0 | 0 | 1.16 |
| BLA | gene expression | 0 | 0 | 1.15 |
| BLA | isoform ratio | 0 | 0 | 1.17 |
| BLA | intron excision ratio | 0 | 0 | 1.17 |
| BLA | mRNA stability | 0 | 0 | 1.09 |
| Brain | alternative polyA | 0 | 0 | 1.12 |
| Brain | alternative TSS | 0 | 0 | 1.14 |
| Brain | gene expression | 0 | 0 | 1.12 |
| Brain | isoform ratio | 0 | 0 | 1.11 |
| Brain | intron excision ratio | 0 | 0 | 1.12 |
| Brain | mRNA stability | 0 | 0 | 1.1 |
| Eye | alternative polyA | 0 | 0 | 1.47 |
| Eye | alternative TSS | 0 | 0 | 1.08 |
| Eye | gene expression | 0 | 0 | 1.23 |
| Eye | isoform ratio | 0 | 0 | 1.06 |
| Eye | intron excision ratio | 0 | 0 | 1.22 |
| Eye | mRNA stability | 0 | 0 | 1.19 |
| IL | alternative polyA | 0 | 0 | 1.18 |
| IL | alternative TSS | 0 | 0 | 1.2 |
| IL | gene expression | 0 | 0 | 1.16 |
| IL | isoform ratio | 0 | 0 | 1.26 |
| IL | intron excision ratio | 0 | 0 | 1.35 |
| IL | mRNA stability | 0 | 0 | 1.07 |
| LHb | alternative polyA | 0 | 0 | 1.18 |
| LHb | alternative TSS | 0 | 0 | 1.48 |
| LHb | gene expression | 0 | 0 | 1.17 |
| LHb | isoform ratio | 0 | 0 | 1.22 |
| LHb | intron excision ratio | 0 | 0 | 1.41 |
| LHb | mRNA stability | 0 | 0 | 1.18 |
| Liver | alternative polyA | 0 | 0 | 1.18 |
| Liver | alternative TSS | 0 | 0 | 1.27 |
| Liver | gene expression | 0 | 0 | 1.15 |
| Liver | isoform ratio | 0 | 0 | 1.19 |
| Liver | intron excision ratio | 0 | 0 | 1.15 |
| Liver | mRNA stability | 0 | 0 | 1.11 |
| NAcc | alternative polyA | 0 | 0 | 1.1 |
| NAcc | alternative TSS | 0 | 0 | 1.22 |
| NAcc | gene expression | 0 | 0 | 1.13 |
| NAcc | isoform ratio | 0 | 0 | 1.17 |
| NAcc | intron excision ratio | 0 | 0 | 1.19 |
| NAcc | mRNA stability | 0 | 0 | 1.12 |
| OFC | alternative polyA | 0 | 0 | 1.19 |
| OFC | alternative TSS | 0 | 0 | 1.07 |
| OFC | gene expression | 0 | 0 | 1.19 |
| OFC | isoform ratio | 0 | 0 | 1.16 |
| OFC | intron excision ratio | 0 | 0 | 1.48 |
| OFC | mRNA stability | 0 | 0 | 1.05 |
| PL | alternative polyA | 0 | 0 | 1.16 |
| PL | alternative TSS | 0 | 0 | 1.11 |
| PL | gene expression | 0 | 0 | 1.13 |
| PL | isoform ratio | 0 | 0 | 1.16 |
| PL | intron excision ratio | 0 | 0 | 1.24 |
| PL | mRNA stability | 0 | 0 | 1.09 |
| pVTA | alternative polyA | 0 | 0 | 1.25 |
| pVTA | alternative TSS | 0 | 0 | 1.18 |
| pVTA | gene expression | 0 | 0 | 1.15 |
| pVTA | isoform ratio | 0 | 0 | 1.21 |
| pVTA | intron excision ratio | 0 | 0 | 1.27 |
| pVTA | mRNA stability | 0 | 0 | 1.09 |
| RMTg | alternative polyA | 0 | 0 | 1.34 |
| RMTg | alternative TSS | 0 | 0 | 1.23 |
| RMTg | gene expression | 0 | 0 | 1.19 |
| RMTg | isoform ratio | 0 | 0 | 1.33 |
| RMTg | intron excision ratio | 0 | 0 | 1.21 |
| RMTg | mRNA stability | 0 | 0 | 1.21 |