Time in novel zone, NPP test
Time in novel zone during novelty place preference test for males and females [sec]
Tags: Behavior · Motivation · Novelty
Project: p50_shelly_flagel_2014
1 locus · 1 gene with independent associations · 1 total associated gene
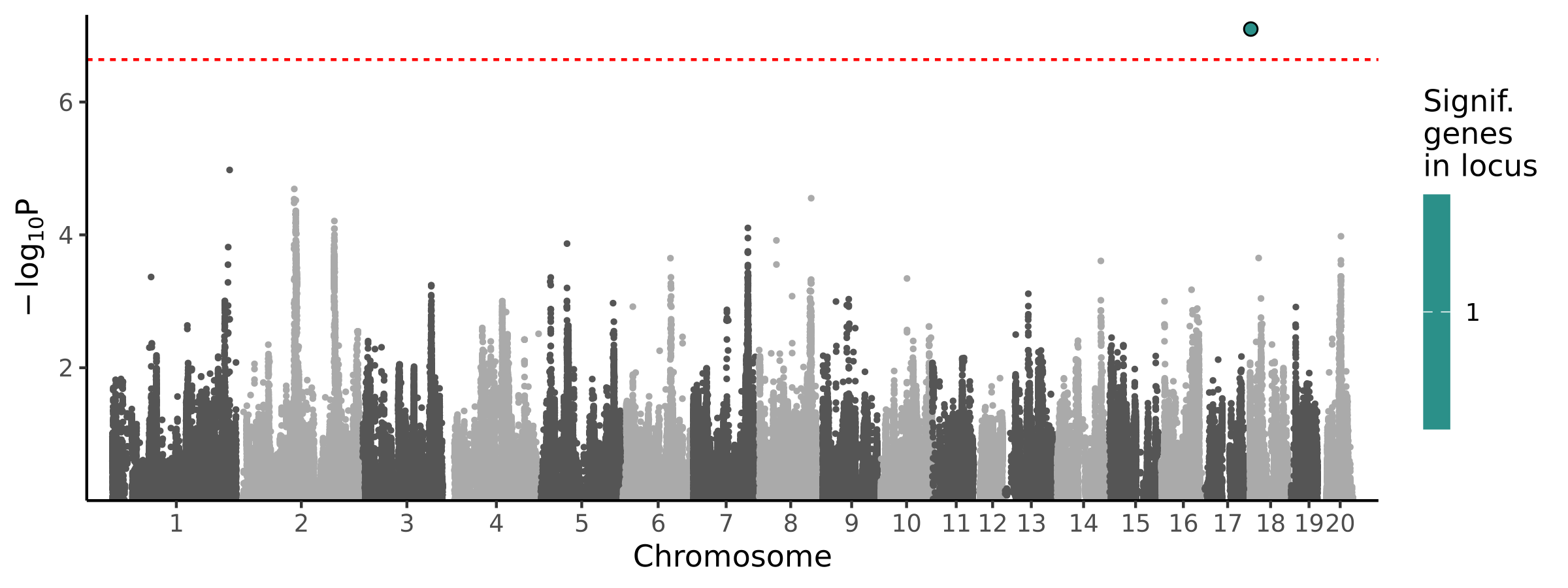
Significant Loci
| # | Chr | Start pos | End pos | # assoc genes | # joint models | Best TWAS P | Best GWAS P | Cond GWAS P | Joint genes |
|---|---|---|---|---|---|---|---|---|---|
| 1 | chr18 | 969654 | 2367828 | 1 | 1 | 7.97e-08 | 1.74e-02 | 1.74e-04 | LOC680026 |
Pleiotropic Associations
Associations by panel
| Tissue | RNA modality | # hits | % hits/tests | Avg chisq |
|---|---|---|---|---|
| Adipose | alternative polyA | 0 | 0 | 1.04 |
| Adipose | alternative TSS | 0 | 0 | 1.08 |
| Adipose | gene expression | 1 | 0 | 1.02 |
| Adipose | isoform ratio | 0 | 0 | 1.03 |
| Adipose | intron excision ratio | 0 | 0 | 0.98 |
| Adipose | mRNA stability | 0 | 0 | 1.09 |
| BLA | alternative polyA | 0 | 0 | 1.01 |
| BLA | alternative TSS | 0 | 0 | 1.02 |
| BLA | gene expression | 0 | 0 | 1.04 |
| BLA | isoform ratio | 0 | 0 | 1.03 |
| BLA | intron excision ratio | 0 | 0 | 0.99 |
| BLA | mRNA stability | 0 | 0 | 1.02 |
| Brain | alternative polyA | 0 | 0 | 1.02 |
| Brain | alternative TSS | 0 | 0 | 1.05 |
| Brain | gene expression | 0 | 0 | 1.04 |
| Brain | isoform ratio | 0 | 0 | 0.99 |
| Brain | intron excision ratio | 0 | 0 | 1.06 |
| Brain | mRNA stability | 0 | 0 | 1.06 |
| Eye | alternative polyA | 0 | 0 | 1.01 |
| Eye | alternative TSS | 0 | 0 | 1.08 |
| Eye | gene expression | 0 | 0 | 1.07 |
| Eye | isoform ratio | 0 | 0 | 1.01 |
| Eye | intron excision ratio | 0 | 0 | 0.95 |
| Eye | mRNA stability | 0 | 0 | 1.2 |
| IL | alternative polyA | 0 | 0 | 0.91 |
| IL | alternative TSS | 0 | 0 | 1.02 |
| IL | gene expression | 0 | 0 | 1.02 |
| IL | isoform ratio | 0 | 0 | 0.95 |
| IL | intron excision ratio | 0 | 0 | 0.92 |
| IL | mRNA stability | 0 | 0 | 1.02 |
| LHb | alternative polyA | 0 | 0 | 0.99 |
| LHb | alternative TSS | 0 | 0 | 0.99 |
| LHb | gene expression | 0 | 0 | 1.06 |
| LHb | isoform ratio | 0 | 0 | 1.04 |
| LHb | intron excision ratio | 0 | 0 | 1.02 |
| LHb | mRNA stability | 0 | 0 | 1.11 |
| Liver | alternative polyA | 0 | 0 | 1.06 |
| Liver | alternative TSS | 0 | 0 | 1.05 |
| Liver | gene expression | 0 | 0 | 1.04 |
| Liver | isoform ratio | 0 | 0 | 1.03 |
| Liver | intron excision ratio | 0 | 0 | 1.03 |
| Liver | mRNA stability | 0 | 0 | 1.05 |
| NAcc | alternative polyA | 0 | 0 | 1.03 |
| NAcc | alternative TSS | 0 | 0 | 1.04 |
| NAcc | gene expression | 0 | 0 | 1.03 |
| NAcc | isoform ratio | 0 | 0 | 1.02 |
| NAcc | intron excision ratio | 0 | 0 | 1.06 |
| NAcc | mRNA stability | 0 | 0 | 1.04 |
| OFC | alternative polyA | 0 | 0 | 0.92 |
| OFC | alternative TSS | 0 | 0 | 1.07 |
| OFC | gene expression | 0 | 0 | 1.03 |
| OFC | isoform ratio | 0 | 0 | 1.01 |
| OFC | intron excision ratio | 0 | 0 | 0.97 |
| OFC | mRNA stability | 0 | 0 | 1.07 |
| PL | alternative polyA | 0 | 0 | 0.98 |
| PL | alternative TSS | 0 | 0 | 1.02 |
| PL | gene expression | 0 | 0 | 1.02 |
| PL | isoform ratio | 0 | 0 | 1.01 |
| PL | intron excision ratio | 0 | 0 | 1.03 |
| PL | mRNA stability | 0 | 0 | 1.05 |
| pVTA | alternative polyA | 0 | 0 | 1.04 |
| pVTA | alternative TSS | 0 | 0 | 1.02 |
| pVTA | gene expression | 0 | 0 | 1.04 |
| pVTA | isoform ratio | 0 | 0 | 1.05 |
| pVTA | intron excision ratio | 0 | 0 | 1.01 |
| pVTA | mRNA stability | 0 | 0 | 1 |
| RMTg | alternative polyA | 0 | 0 | 0.95 |
| RMTg | alternative TSS | 0 | 0 | 1.05 |
| RMTg | gene expression | 0 | 0 | 1.1 |
| RMTg | isoform ratio | 0 | 0 | 1.01 |
| RMTg | intron excision ratio | 0 | 0 | 1.09 |
| RMTg | mRNA stability | 0 | 0 | 1.07 |