Pavlov. Cond. lever latency
Pavlovian Conditioned Approach, Sign-tracking, average lever latency on days 4 and 5 of testing
Tags: Behavior · Pavlovian conditioned approach · Conditioned reinforcement
Project: p50_paul_meyer_2014
3 loci · 8 genes with independent associations · 171 total associated genes
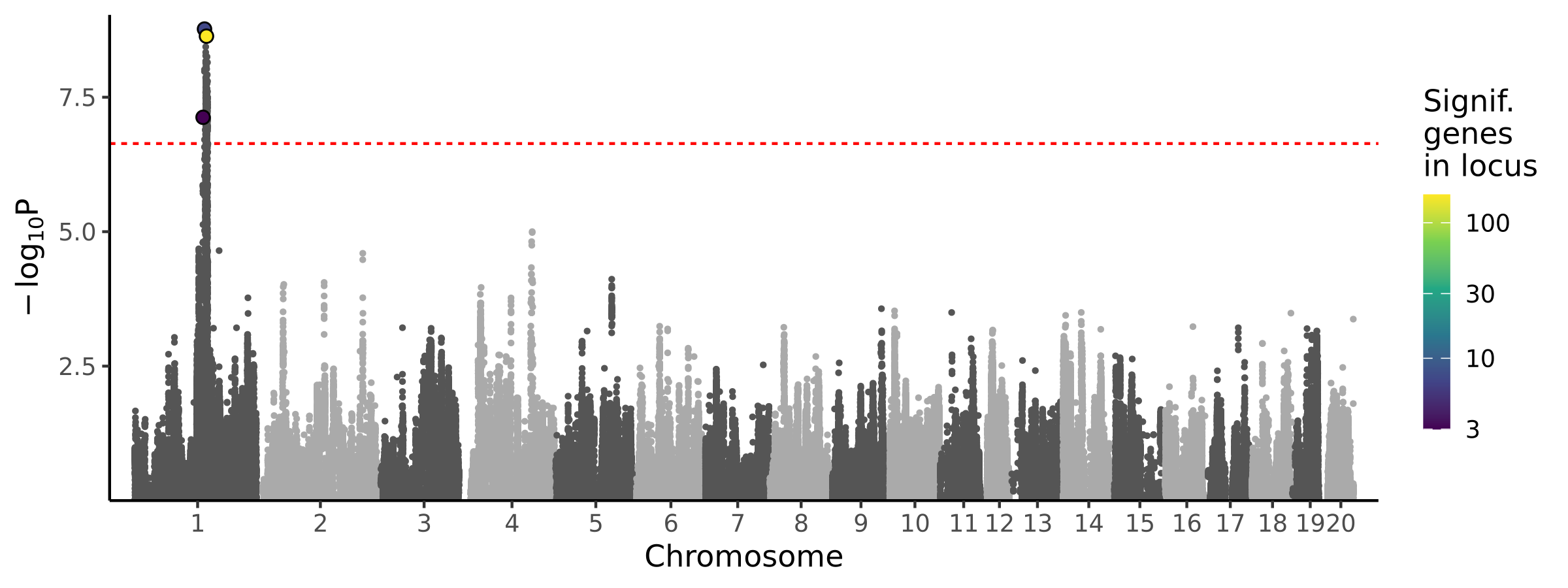
Significant Loci
| # | Chr | Start pos | End pos | # assoc genes | # joint models | Best TWAS P | Best GWAS P | Cond GWAS P | Joint genes |
|---|---|---|---|---|---|---|---|---|---|
| 1 | chr1 | 146106284 | 147631216 | 3 | 1 | 7.47e-08 | 1.22e-06 | 1.00e+00 | Rab30 |
| 2 | chr1 | 149199615 | 150594931 | 7 | 1 | 1.71e-09 | 2.46e-09 | 1.64e-01 | Tenm4 |
| 3 | chr1 | 150731124 | 157355948 | 161 | 6 | 2.32e-09 | 5.61e-09 | 1.00e+00 | Capn5 Emsy LOC120099889 Pak1 Slco2b1 Wnt11 |
Pleiotropic Associations
Associations by panel
| Tissue | RNA modality | # hits | % hits/tests | Avg chisq |
|---|---|---|---|---|
| Adipose | alternative polyA | 10 | 0.3 | 1.36 |
| Adipose | alternative TSS | 15 | 0.4 | 1.43 |
| Adipose | gene expression | 13 | 0.1 | 1.36 |
| Adipose | isoform ratio | 13 | 0.3 | 1.4 |
| Adipose | intron excision ratio | 3 | 0.1 | 1.2 |
| Adipose | mRNA stability | 11 | 0.2 | 1.39 |
| BLA | alternative polyA | 3 | 0.1 | 1.36 |
| BLA | alternative TSS | 7 | 0.4 | 1.39 |
| BLA | gene expression | 13 | 0.2 | 1.37 |
| BLA | isoform ratio | 1 | 0 | 1.4 |
| BLA | intron excision ratio | 3 | 0.1 | 1.32 |
| BLA | mRNA stability | 4 | 0.2 | 1.36 |
| Brain | alternative polyA | 2 | 0.1 | 1.32 |
| Brain | alternative TSS | 10 | 0.2 | 1.34 |
| Brain | gene expression | 13 | 0.1 | 1.35 |
| Brain | isoform ratio | 6 | 0.1 | 1.35 |
| Brain | intron excision ratio | 9 | 0.2 | 1.3 |
| Brain | mRNA stability | 9 | 0.2 | 1.35 |
| Eye | alternative polyA | 0 | 0 | 1.32 |
| Eye | alternative TSS | 0 | 0 | 1.33 |
| Eye | gene expression | 3 | 0.2 | 1.46 |
| Eye | isoform ratio | 0 | 0 | 1.22 |
| Eye | intron excision ratio | 1 | 0.1 | 1.38 |
| Eye | mRNA stability | 0 | 0 | 1.53 |
| IL | alternative polyA | 0 | 0 | 1.27 |
| IL | alternative TSS | 0 | 0 | 1.13 |
| IL | gene expression | 5 | 0.1 | 1.38 |
| IL | isoform ratio | 0 | 0 | 1.36 |
| IL | intron excision ratio | 1 | 0.1 | 1.31 |
| IL | mRNA stability | 1 | 0.1 | 1.34 |
| LHb | alternative polyA | 0 | 0 | 1.39 |
| LHb | alternative TSS | 0 | 0 | 1.34 |
| LHb | gene expression | 2 | 0.1 | 1.34 |
| LHb | isoform ratio | 1 | 0.1 | 1.28 |
| LHb | intron excision ratio | 0 | 0 | 1.25 |
| LHb | mRNA stability | 0 | 0 | 1.26 |
| Liver | alternative polyA | 1 | 0 | 1.26 |
| Liver | alternative TSS | 3 | 0.1 | 1.36 |
| Liver | gene expression | 18 | 0.2 | 1.39 |
| Liver | isoform ratio | 5 | 0.1 | 1.35 |
| Liver | intron excision ratio | 8 | 0.2 | 1.38 |
| Liver | mRNA stability | 4 | 0.1 | 1.37 |
| NAcc | alternative polyA | 2 | 0.1 | 1.38 |
| NAcc | alternative TSS | 8 | 0.3 | 1.32 |
| NAcc | gene expression | 11 | 0.1 | 1.37 |
| NAcc | isoform ratio | 4 | 0.1 | 1.36 |
| NAcc | intron excision ratio | 4 | 0.1 | 1.3 |
| NAcc | mRNA stability | 7 | 0.2 | 1.37 |
| OFC | alternative polyA | 2 | 0.2 | 1.42 |
| OFC | alternative TSS | 0 | 0 | 1.17 |
| OFC | gene expression | 2 | 0 | 1.32 |
| OFC | isoform ratio | 2 | 0.1 | 1.41 |
| OFC | intron excision ratio | 0 | 0 | 1.24 |
| OFC | mRNA stability | 1 | 0.1 | 1.38 |
| PL | alternative polyA | 9 | 0.3 | 1.47 |
| PL | alternative TSS | 3 | 0.1 | 1.34 |
| PL | gene expression | 15 | 0.2 | 1.36 |
| PL | isoform ratio | 4 | 0.1 | 1.33 |
| PL | intron excision ratio | 6 | 0.1 | 1.31 |
| PL | mRNA stability | 5 | 0.1 | 1.37 |
| pVTA | alternative polyA | 0 | 0 | 1.47 |
| pVTA | alternative TSS | 3 | 0.2 | 1.32 |
| pVTA | gene expression | 6 | 0.1 | 1.38 |
| pVTA | isoform ratio | 4 | 0.2 | 1.32 |
| pVTA | intron excision ratio | 4 | 0.1 | 1.3 |
| pVTA | mRNA stability | 3 | 0.2 | 1.35 |
| RMTg | alternative polyA | 0 | 0 | 1.18 |
| RMTg | alternative TSS | 0 | 0 | 1.18 |
| RMTg | gene expression | 0 | 0 | 1.39 |
| RMTg | isoform ratio | 0 | 0 | 1.09 |
| RMTg | intron excision ratio | 0 | 0 | 1.33 |
| RMTg | mRNA stability | 0 | 0 | 1.41 |