Light reinforcement 2
Light reinforcement, proportion of active snout-poke responses for 5-sec light onset to the total active and inactive responses in the last three 18-minute sessions during the light reinforcement phase for males and females [proportion]
Tags: Behavior · Light reinforcement
Project: p50_david_dietz
2 loci · 2 genes with independent associations · 11 total associated genes
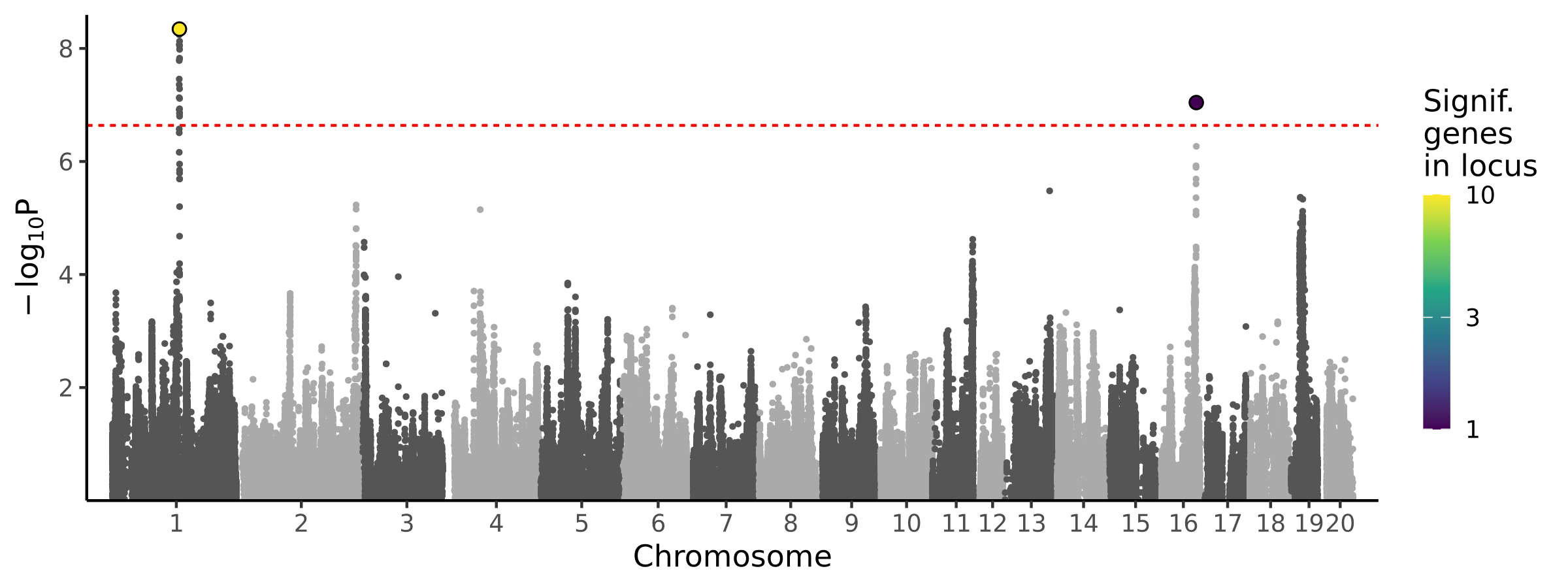
Significant Loci
| # | Chr | Start pos | End pos | # assoc genes | # joint models | Best TWAS P | Best GWAS P | Cond GWAS P | Joint genes |
|---|---|---|---|---|---|---|---|---|---|
| 1 | chr1 | 140610199 | 142727457 | 10 | 1 | 4.54e-09 | 1.24e-09 | 1.00e+00 | Ctsc |
| 2 | chr16 | 72043796 | 73442457 | 1 | 1 | 9.08e-08 | 7.44e-07 | 5.59e-01 | LOC120097408 |
Pleiotropic Associations
Associations by panel
| Tissue | RNA modality | # hits | % hits/tests | Avg chisq |
|---|---|---|---|---|
| Adipose | alternative polyA | 0 | 0 | 1.27 |
| Adipose | alternative TSS | 0 | 0 | 1.33 |
| Adipose | gene expression | 0 | 0 | 1.32 |
| Adipose | isoform ratio | 0 | 0 | 1.29 |
| Adipose | intron excision ratio | 2 | 0.1 | 1.29 |
| Adipose | mRNA stability | 0 | 0 | 1.28 |
| BLA | alternative polyA | 0 | 0 | 1.49 |
| BLA | alternative TSS | 0 | 0 | 1.27 |
| BLA | gene expression | 2 | 0 | 1.33 |
| BLA | isoform ratio | 2 | 0.1 | 1.37 |
| BLA | intron excision ratio | 0 | 0 | 1.35 |
| BLA | mRNA stability | 0 | 0 | 1.31 |
| Brain | alternative polyA | 2 | 0.1 | 1.4 |
| Brain | alternative TSS | 0 | 0 | 1.26 |
| Brain | gene expression | 1 | 0 | 1.34 |
| Brain | isoform ratio | 3 | 0.1 | 1.36 |
| Brain | intron excision ratio | 0 | 0 | 1.41 |
| Brain | mRNA stability | 1 | 0 | 1.34 |
| Eye | alternative polyA | 0 | 0 | 1.19 |
| Eye | alternative TSS | 0 | 0 | 1.44 |
| Eye | gene expression | 1 | 0.1 | 1.3 |
| Eye | isoform ratio | 0 | 0 | 1.36 |
| Eye | intron excision ratio | 0 | 0 | 1.34 |
| Eye | mRNA stability | 0 | 0 | 1.46 |
| IL | alternative polyA | 0 | 0 | 1.36 |
| IL | alternative TSS | 0 | 0 | 1.34 |
| IL | gene expression | 1 | 0 | 1.36 |
| IL | isoform ratio | 0 | 0 | 1.41 |
| IL | intron excision ratio | 0 | 0 | 1.29 |
| IL | mRNA stability | 0 | 0 | 1.31 |
| LHb | alternative polyA | 0 | 0 | 1.31 |
| LHb | alternative TSS | 0 | 0 | 1.28 |
| LHb | gene expression | 0 | 0 | 1.3 |
| LHb | isoform ratio | 0 | 0 | 1.27 |
| LHb | intron excision ratio | 0 | 0 | 1.38 |
| LHb | mRNA stability | 0 | 0 | 1.36 |
| Liver | alternative polyA | 0 | 0 | 1.36 |
| Liver | alternative TSS | 0 | 0 | 1.3 |
| Liver | gene expression | 1 | 0 | 1.3 |
| Liver | isoform ratio | 0 | 0 | 1.29 |
| Liver | intron excision ratio | 0 | 0 | 1.31 |
| Liver | mRNA stability | 0 | 0 | 1.29 |
| NAcc | alternative polyA | 0 | 0 | 1.34 |
| NAcc | alternative TSS | 0 | 0 | 1.33 |
| NAcc | gene expression | 2 | 0 | 1.33 |
| NAcc | isoform ratio | 1 | 0 | 1.42 |
| NAcc | intron excision ratio | 0 | 0 | 1.43 |
| NAcc | mRNA stability | 0 | 0 | 1.39 |
| OFC | alternative polyA | 0 | 0 | 1.36 |
| OFC | alternative TSS | 0 | 0 | 1.34 |
| OFC | gene expression | 0 | 0 | 1.31 |
| OFC | isoform ratio | 0 | 0 | 1.44 |
| OFC | intron excision ratio | 0 | 0 | 1.33 |
| OFC | mRNA stability | 0 | 0 | 1.32 |
| PL | alternative polyA | 0 | 0 | 1.39 |
| PL | alternative TSS | 0 | 0 | 1.36 |
| PL | gene expression | 1 | 0 | 1.33 |
| PL | isoform ratio | 0 | 0 | 1.38 |
| PL | intron excision ratio | 0 | 0 | 1.41 |
| PL | mRNA stability | 0 | 0 | 1.36 |
| pVTA | alternative polyA | 0 | 0 | 1.28 |
| pVTA | alternative TSS | 0 | 0 | 1.33 |
| pVTA | gene expression | 0 | 0 | 1.33 |
| pVTA | isoform ratio | 0 | 0 | 1.3 |
| pVTA | intron excision ratio | 0 | 0 | 1.37 |
| pVTA | mRNA stability | 0 | 0 | 1.35 |
| RMTg | alternative polyA | 0 | 0 | 1.2 |
| RMTg | alternative TSS | 0 | 0 | 1.24 |
| RMTg | gene expression | 0 | 0 | 1.41 |
| RMTg | isoform ratio | 0 | 0 | 1.42 |
| RMTg | intron excision ratio | 0 | 0 | 1.35 |
| RMTg | mRNA stability | 0 | 0 | 1.3 |