Social time
Social time indicated by average time that the rat spent with its head in the observation port providing access to the stimulus rat while the sliding door was open in the last three 18-minute sessions for males and females [cs]
Tags: Behavior · Social reinforcement
Project: p50_david_dietz
1 locus · 1 gene with independent associations · 1 total associated gene
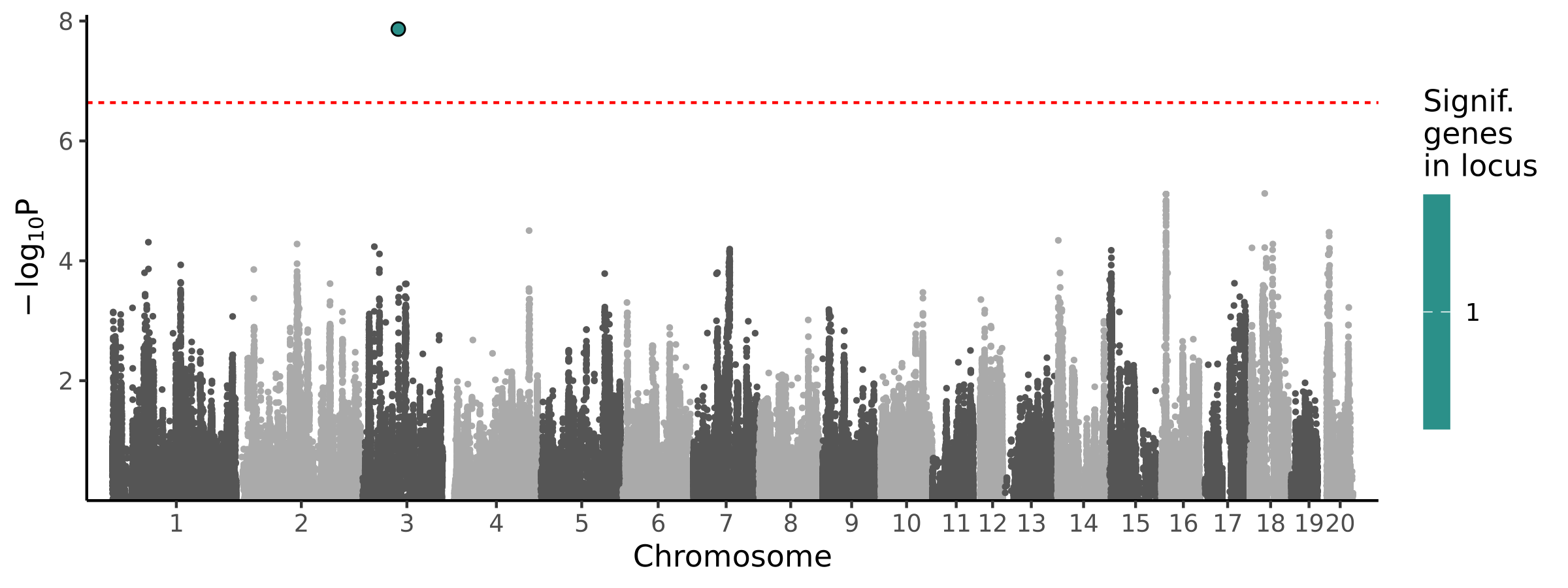
Significant Loci
| # | Chr | Start pos | End pos | # assoc genes | # joint models | Best TWAS P | Best GWAS P | Cond GWAS P | Joint genes |
|---|---|---|---|---|---|---|---|---|---|
| 1 | chr3 | 76033161 | 77431053 | 1 | 1 | 1.36e-08 | 4.87e-03 | 1.6e-05 | Fnbp4 |
Pleiotropic Associations
Associations by panel
| Tissue | RNA modality | # hits | % hits/tests | Avg chisq |
|---|---|---|---|---|
| Adipose | alternative polyA | 0 | 0 | 1.27 |
| Adipose | alternative TSS | 0 | 0 | 1.3 |
| Adipose | gene expression | 0 | 0 | 1.26 |
| Adipose | isoform ratio | 1 | 0 | 1.35 |
| Adipose | intron excision ratio | 0 | 0 | 1.46 |
| Adipose | mRNA stability | 0 | 0 | 1.29 |
| BLA | alternative polyA | 0 | 0 | 1.33 |
| BLA | alternative TSS | 0 | 0 | 1.29 |
| BLA | gene expression | 0 | 0 | 1.33 |
| BLA | isoform ratio | 0 | 0 | 1.3 |
| BLA | intron excision ratio | 0 | 0 | 1.41 |
| BLA | mRNA stability | 0 | 0 | 1.29 |
| Brain | alternative polyA | 0 | 0 | 1.35 |
| Brain | alternative TSS | 0 | 0 | 1.29 |
| Brain | gene expression | 0 | 0 | 1.3 |
| Brain | isoform ratio | 0 | 0 | 1.32 |
| Brain | intron excision ratio | 0 | 0 | 1.4 |
| Brain | mRNA stability | 0 | 0 | 1.31 |
| Eye | alternative polyA | 0 | 0 | 1.71 |
| Eye | alternative TSS | 0 | 0 | 1.51 |
| Eye | gene expression | 0 | 0 | 1.39 |
| Eye | isoform ratio | 0 | 0 | 1.32 |
| Eye | intron excision ratio | 0 | 0 | 1.53 |
| Eye | mRNA stability | 0 | 0 | 1.56 |
| IL | alternative polyA | 0 | 0 | 1.46 |
| IL | alternative TSS | 0 | 0 | 1.27 |
| IL | gene expression | 0 | 0 | 1.35 |
| IL | isoform ratio | 0 | 0 | 1.37 |
| IL | intron excision ratio | 0 | 0 | 1.26 |
| IL | mRNA stability | 0 | 0 | 1.25 |
| LHb | alternative polyA | 0 | 0 | 1.26 |
| LHb | alternative TSS | 0 | 0 | 1.35 |
| LHb | gene expression | 0 | 0 | 1.32 |
| LHb | isoform ratio | 0 | 0 | 1.31 |
| LHb | intron excision ratio | 0 | 0 | 1.5 |
| LHb | mRNA stability | 0 | 0 | 1.29 |
| Liver | alternative polyA | 0 | 0 | 1.34 |
| Liver | alternative TSS | 0 | 0 | 1.26 |
| Liver | gene expression | 0 | 0 | 1.27 |
| Liver | isoform ratio | 0 | 0 | 1.3 |
| Liver | intron excision ratio | 0 | 0 | 1.38 |
| Liver | mRNA stability | 0 | 0 | 1.27 |
| NAcc | alternative polyA | 0 | 0 | 1.36 |
| NAcc | alternative TSS | 0 | 0 | 1.29 |
| NAcc | gene expression | 0 | 0 | 1.32 |
| NAcc | isoform ratio | 0 | 0 | 1.3 |
| NAcc | intron excision ratio | 0 | 0 | 1.32 |
| NAcc | mRNA stability | 0 | 0 | 1.3 |
| OFC | alternative polyA | 0 | 0 | 1.42 |
| OFC | alternative TSS | 0 | 0 | 1.2 |
| OFC | gene expression | 0 | 0 | 1.31 |
| OFC | isoform ratio | 0 | 0 | 1.35 |
| OFC | intron excision ratio | 0 | 0 | 1.37 |
| OFC | mRNA stability | 0 | 0 | 1.35 |
| PL | alternative polyA | 0 | 0 | 1.35 |
| PL | alternative TSS | 0 | 0 | 1.25 |
| PL | gene expression | 0 | 0 | 1.3 |
| PL | isoform ratio | 0 | 0 | 1.31 |
| PL | intron excision ratio | 0 | 0 | 1.31 |
| PL | mRNA stability | 0 | 0 | 1.32 |
| pVTA | alternative polyA | 0 | 0 | 1.34 |
| pVTA | alternative TSS | 0 | 0 | 1.24 |
| pVTA | gene expression | 0 | 0 | 1.36 |
| pVTA | isoform ratio | 0 | 0 | 1.28 |
| pVTA | intron excision ratio | 0 | 0 | 1.37 |
| pVTA | mRNA stability | 0 | 0 | 1.37 |
| RMTg | alternative polyA | 0 | 0 | 1.44 |
| RMTg | alternative TSS | 0 | 0 | 1.23 |
| RMTg | gene expression | 0 | 0 | 1.4 |
| RMTg | isoform ratio | 0 | 0 | 1.33 |
| RMTg | intron excision ratio | 0 | 0 | 1.6 |
| RMTg | mRNA stability | 0 | 0 | 1.4 |